The most important genetic association in rheumatoid arthritis (RA) is presented with some alleles from the HLA-DRB1 gene that encode the shared epitope (SE).
ObjectivesTo apply the SE classification methods of Gregersen, de Vries, Raychaudhuri, Mattey, and Tezenas du Montcel in a group of Colombian patients with RA and determine the most common HLA-DRB1 alleles in the population.
MethodsRA diagnosis, genetic study of the HLA-DRB1 region using Luminex technology in 50 RA and 50 healthy subjects. For the classification analysis, Fisher's exact test and chi-squared test were applied. Tables were created to count the RA-related alleles. We used odds ratio to determine the risk between the presence of the shared epitope (SE) and anti-cyclic citrullinated peptides (Anti-CCP).
ResultsGregersen and de Vries methods were suitable for the characterization of RA in this population (p = .006). The most prevalent HLA-DRB1 alleles in the RA group were 14:02, 04:04, 08:02, 04:05, and 10:01. High frequencies of the 07:01, 03:01, 13:02, 01:02, and 12:01 HLA-DRB1 alleles were found in the healthy population. HLA-DRB1 alleles with similar distribution in both populations were 04:07, 15:01, 11:01, 16:02, and 01:01. A high frequency of SE + was observed in Anti-CCP + individuals (63.15%); however, this was not statistically significant [OR 2.4 (.63–9.01); p = .19].
ConclusionThe SE classification methods of Gregersen and de Vries were adequate in characterizing RA in a Colombian population group. An equivalence of 100% was verified between the susceptibility alleles defined by de Vries and the alleles assigned as SE according to Gregersen.
La asociación genética más importante en artritis reumatoide (AR) se presenta con algunos alelos del gen HLA DRB1 que codifican el epítope compartido (EC).
ObjetivosAplicar los métodos de clasificación de EC de Gregersen, de Vries, Raychaudhuri, Mattey y Tezenas du Montcel en un grupo de pacientes colombianos con AR, y determinar los alelos HLA DRB1 más frecuentes en esta población.
MétodosDiagnóstico para AR, estudio genético de la región HLA DRB1 por tecnología Luminex de 50 sujetos AR y 50 sanos. Para análisis comparativos de clasificaciones EC, se aplicaron las pruebas test exacto de Fisher y Chi cuadrado y se realizaron tablas de conteos para los alelos relacionados con AR. Se estimó la razón de Odds para determinar el riesgo entre la presencia de EC y los anticuerpos antipéptidos cíclicos citrulinados (anti-PCC).
ResultadosLos métodos de Gregersen y de Vries fueron adecuados para la caracterización de AR en esta población (p = 0,006). Los alelos HLA DRB1 más prevalentes en el grupo AR fueron 14:02, 04:04, 08:02, 04:05 y 10:01. Se encontraron altas frecuencias de los alelos HLA DRB1 07:01, 03:01, 13:02, 01:02 y 12:01 en población sana. Alelos HLA DRB1 con distribución similar en ambas poblaciones fueron: 04:07, 15:01, 11:01, 16:02 y 01:01. Se observó alta frecuencia de individuos EC + en el grupo AR Anti-PCC+ (63,15%); no obstante, sin asociación estadística [OR 2,4 (0,63−9,01), p = 0,19]
ConclusiónLos métodos de clasificación para EC de Gregersen y de Vries fueron adecuados caracterizando AR en un grupo de población colombiana. Se corroboró equivalencia del 100% entre los alelos de susceptibilidad definidos por de Vries y los alelos asignados como EC según Gregersen.
The HLA DRB1 locus is the most significant genetic risk factor associated with rheumatoid arthritis (RA).1,2 Studies on this relationship date back from the 1970s, in which it was observed that the HLA DR4 subtype had a higher frequency in patients with RA, compared with the control population.3 Other HLA DR alleles, for example, HLA DR14 in Native Americans4 or HLA DR1 and DR10 in Caucasian populations,5 are also related with the disease. The sequence analysis of the HLA DRB1 alleles associated with RA showed that the majority of patients with RA share conserved amino acid sequences in the third hypervariable region (HV3) of the HLA DRβ molecule, which have been named shared epitopes (SE).6,7 The alleles that code for the SE are associated with susceptibility for RA6–9 and also with the severity of the disease10,11; in addition, they have been associated with the production of anti-cyclic citrullinated peptide (anti-CCP) antibodies.12,13 Although there is general agreement on the predisposition of the alleles with SE for RA, not all entail the same risk,5,7,14,15 there are also discrepancies related to the precise alleles that determine this susceptibility.12,16
A great number of modifications have been proposed regarding the classification of the SE in different populations analyzed, in relation to variations in the positions of the aminoacids within the HLA DRβ chain. The objective of this study was to determine the HLA DRB1 alleles in a group of Colombian individuals with and without RA, and correlate the classification methods for SE of Gregersen et al.,6 Vrieset al.,5 Raychaudhuri et al.,14 Mattey et al.7 and Tezenas du Montcel et al.15 in this population.
Classification of Gregersen et alThe classical classification of SE is based on conserved sequences of three variants of homologous aminoacids at positions 70–74 of the HLA DRβ1 molecule.6,7 These sequences correspond to the aminoacids QKRAA (alleles 04:01, 04:11, 04:09), QRRAA (alleles 01:01, 01:02, 01:04, 01:05, 04:04, 04:05, 04:08, 14:02) and RRRAA (alleles 10:01 and 10:02).
Classification of Tezenas du Montcel et alTezenas du Montcel et al.15 suggested that the risk represented by the presence of the RAA sequence in the positions 72–74 is modulated by the aminoacids in the positions 70 and 71 of the HLA DRβ1 chain. The study was conducted in French patients with RA, and the alleles were divided into two groups according with the presence or absence of the RAA sequence in the positions 72–74, represented as S or X alleles, respectively. The S alleles were divided subsequently according to the aminoacid in the position 71: S1 for ARAA and ERAA, S2 for KRAA and S3 for RRAA. The S3 alleles were further subdivided according to the aminoacid in the position 70, in such a way that S3D corresponded to the DRRAA sequence and S3P to the QRRAA or RRRAA sequences. Comparisons between alleles showed that the risk associated with S1, S3D, and X did not differ significantly and that these alleles represented low risk, which is why they were grouped as a single allele named L. The following hierarchy of risk was established: significatively higher for S2 with respect to S3P (p < 0.002), which in turn was higher than that assigned to L. The genotype with the highest risk was S2/S3P; this risk was 6.6 times higher with respect to the L/L genotype, followed by S2/S2, S3P/S3P, S2/L and S3P/L.15
Classification of De Vries et alStudies in Caucasian population considered the SE according to the positions of aminoacids 67, 70, 71, 73 and 74, corresponding to the sequences LQKAA (DRB1*04:01 or 04:09), LQRAA (DRB1*01:01, 01:02, 04:04, 04:05, 04:08, 14:02) or LRRAA (DRB1*10:01), and called them susceptibility (S) alleles. In addition, they classified the alleles encoding isoleucine in the position 67 or aspartic acid in the position 70 HLA DRβ1 as protective (P) and all other alleles as neutral (N).5
Classification of Mattey et alIn studies conducted in populations of the United Kingdom and Spain, the HLA DRB1 alleles were stratified into five groups, depending on the aminoacid at position 70 and whether or not they were part of the classical sequence of the SE, and the following risk hierarchies were established: Q70SE+ > Q70SE+ > R70SE+ > R70SE+ > D70SE+.7
Classification of Raychaudhuri et alGenome-wide single nucleotide polymorphism (SNP) data, from a GWAS meta-analysis of RA for HLA-A, B, C, DPA1, DPB1, DQA1, DQB1, and DRB1 in the major histocompatibility complex (MHC) were used in this case. The haplotype analyzes revealed that amino acids at positions 11 (or 13) 71 and 74 HLA DRβ1 were associated with risk for RA, and 16 risk haplotypes were defined.14
Patients and methodsThe patients included in the study attended the outpatient clinic at the Central Military Hospital (Hospital Militar Central), located in Bogotá (Colombia), during the period between May 2012 and May 2014, and approved their participation in the study by signing an informed consent. The ethics committees of the participating institutions, in accordance with the principles of the World Medical Association set forth in its Declaration of Helsinki, approved the research: Universidad Nacional de Colombia (24/10/2013−62), Hospital Militar Central (2014–4337). The study included 50 individuals with a diagnosis of RA, according to the American College of Rheumatology classification criteria of 198717 or 2010,18 and 50 individuals without RA who did not meet these diagnostic criteria and did not have a family history of autoimmunity. With respect to ancestry, for the entire population that belonged to this investigation, it was taken into account that at least three generational lines of the participating individuals had been born in Colombian territory; the sample was considered constituted by Colombian mestizos. The healthy population was selected in the same hospital, at the workplace, or they were neighbors of the patients, with similar environmental and socioeconomic conditions, and were matched by age and sex, except for one male individual in the group without RA.
Exclusion criteriaThe participants in this study were also part of an alternate investigation (results are in publication process), and the following exclusion criteria were taken into account: current infection or diagnosis of neoplasia, presence of other autoimmune diseases, treatment with antibiotics in the last three months, having less than six teeth in the mouth, periodontal or orthodontic treatment in the last six months, pregnant or lactating women, current or past smoking habit.
Anti-cyclic citrullinated peptide antibodies IgG/IgAThe Quanta Lite CCP3.1® IgG/IgA ELISA kit was used according to the manufacturer’s instructions. The final result was obtained in ELISA units/mL, and < 20 IU was considered a negative value.
Characterization of HLA DRB1 allelesDNA extraction was performed with the Salting Out method, CorpoGen® kit. The typification of HLA DRB1 alleles was performed in a Luminex® 100/200 platform, using the kit Inmucor, Lifecodes® HLA-DRB1 SSO Typing Kit, 628923, Stamford, CT, USA, following the manufacturer’s instructions; data were analyzed using the LifeMatch DNA software.
Statistical analysisThe SPSS® version 20.0 and R version 3.2.2 programs were used, Chi-square or Fisher tests were applied selecting one or the other based on the number of events within the categories for each classification method. Counting tables were made for the alleles related with RA, with healthy individuals and those with similar distribution in the population. The odds ratio was estimated to determine the risk between the presence of SE and anti-CCP.
ResultsClinical characteristics, relationship between anti-cyclic citrullinated peptide antibodies and shared epitopeThe study population consisted of 100 individuals: Colombian men and women no smokers, between 18 and 65 years of age; 50 with a diagnosis of RA and 50 healthy individuals. The presence of anti-CCP was exclusive of the population with RA (p = 0.001), with a positivity of 76%. The distribution of the disease with respect to the gender reflected the higher occurrence of RA in women, being 84% of the female gender (Table 1). A high frequency of SE+ (SE+/SE+ and SE+SE−) individuals was observed in the RA anti-CCP+group (63.15% of the anti-CCP population), taking into account the classical categorization of SE (according to Gregersen et al.). The differences for the anti-CCP+ and anti-CCP− groups related to the presence of the SE were not statistically significant (OR: 2.4 [0.63−9.01]; p = 0.19) (Table 2).
Characteristics of the RA and healthy populations.
| RAn (%)50 (100.00) | Healthy n (%)50 (100.00) | p-value | |
|---|---|---|---|
| Age (average number of years) | 47.83 | 49.06 | NA |
| Women | 42 (84.00%) | 41 (82.00%) | NA |
| Anti-CCP | 38 (76.00%) | 0 | 0.001* |
Anti-CCP, antibodies against cyclic citrullinated peptides; RA, rheumatoid arthritis; NA, not applicable (individuals matched by age and sex, except for one male individual in the group without RA).
Relationship between anti-CCP and SE in individuals with RA according to Gregersen.
| SE | Anti-CCP+n (%)38 (76) | Anti-CCP−n (%)12 (24) | OR (95% CI) | p-value |
|---|---|---|---|---|
| SE+ (%) | 24 (63.15) | 5 (41.66) | 2.4 (0.63−9.01) | 0.19 |
| SE− (%) | 14 (36.84) | 7 (58.33) |
Data available for 50 individuals with RA.
Anti-CCP: antibodies against cyclic citrullinated peptides; RA: Rheumatoid arthritis; SE: shared epitope; SE+ = SE+/SE+ and SE+/SE−. SE− = SE−/SE−; OR: odds ratio.
Since the results were obtained using Luminex® technology (medium resolution), exclusively unambiguous data were included for the analyses, according to the aminoacid sequences established in each of the classification methods, as follows: according to the classic classification of Gregersen et al., information of 97 samples was included (50 RA and 47 healthy; 194 alleles). In the classifications of Vries and Mattey et al. the unambiguous results comprised 50 RA individuals and 45 healthy individuals (190 alleles). For the analysis according to Tezenas du Montcel et al., there were 47 RA individuals and 47 healthy individuals (188 alleles). For the allelic analysis according to Raychaudhuri et al., 97 alleles were obtained unambiguously in the RA population and 92 alleles in the healthy population. Between the classification system for SE proposed by Gregersen et al. and the system of de Vries et al., no differences were observed with respect to the alleles categorized as SE according to Gregersen et al., defined by de Vries et al. as S alleles. The classification systems of Gregersen et al. and de Vries et al. were appropriate for the differentiation between healthy and RA individuals in this population (p = 0.006); the other classification systems did not obtain statistical significance. It was possible to evidence how the classification of Raychaudhuri et al. prevented from making a differentiation between RA/healthy population; regarding the system of Tezenas Du Montcel et al., despite not having statistically significant results, a higher frequency of alleles L was observed in the population without RA (65.95%) (Table 3). According to the classification system of Mattey et al., regarding the relationship between the Q70EC+DR4 and Q70EC+DR1 alleles for RA susceptibility, we found a higher frequency of the DR4 haplotype (data not shown).
Distribution of the shared epitope according to the methods of Gregersen et al., de Vrieset al., Matteyet al., Tezenas du Montcel et al. and Raychaudhuri et al., in healthy Colombian individuals and with rheumatoid arthritis.
| Classification HLA DRβ 1 | RA | Healthy | p-value | ||
|---|---|---|---|---|---|
| n (%) | Allelic frequency RA | n (%) | Allelic frequency healthy | ||
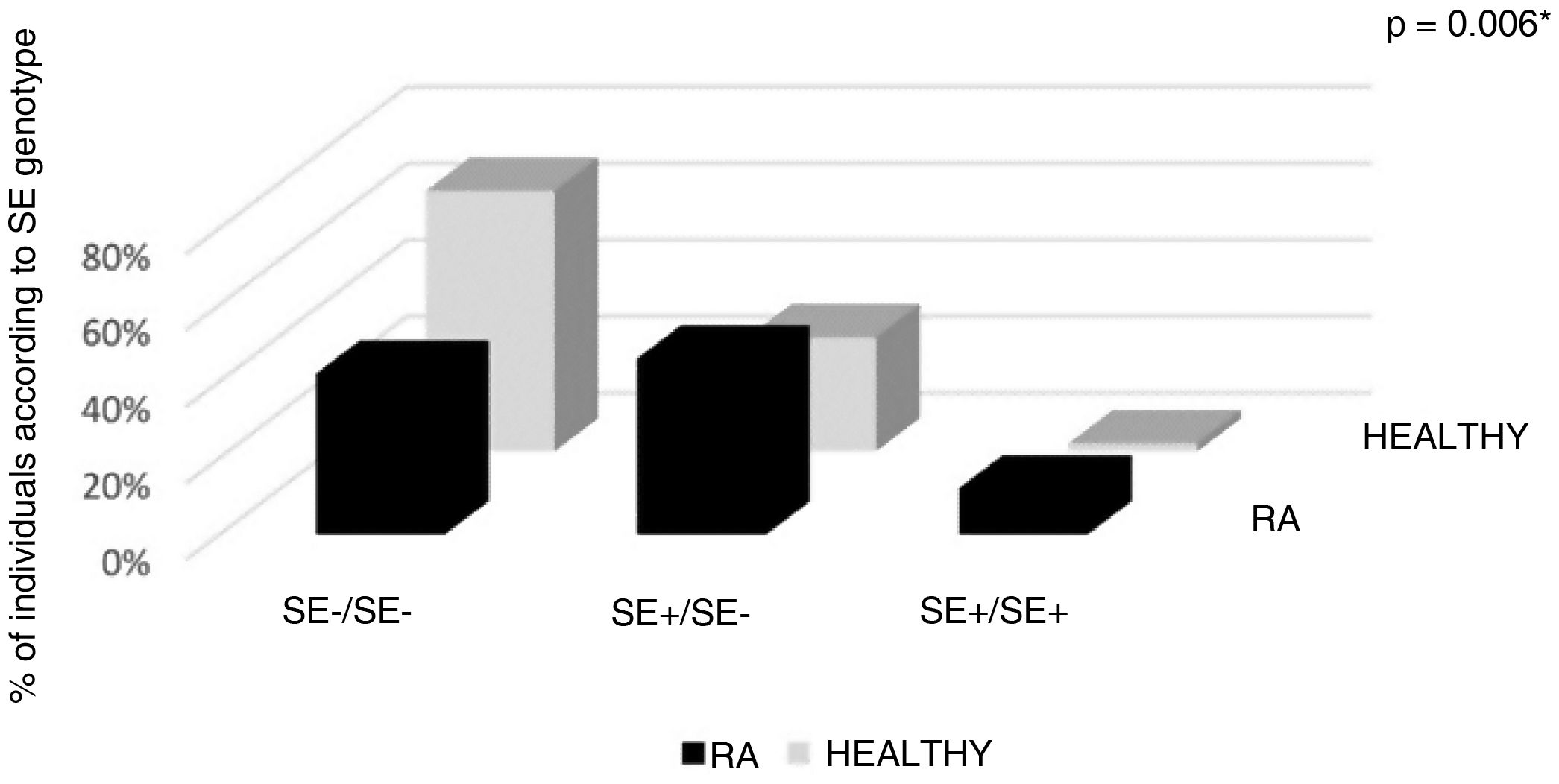
| Gregersen genotype | 50 (100) | 100 | 47 (100) | 94 | *0.006 |
| SE−/SE− | 21 (42) | 42 | 32 (68.1) | 64 | |
| SE+/SE− | 23 (46) | 23(SE+)/23(SE−) | 14 (29.8) | 14(SE+)/14(SE−) | |
| SE+/SE+ | 6 (12) | 12 | 1 (2.1) | 2 | |
| ≥1 SE+ | 29 (58) | 35 | 15 (32) | 16 | |
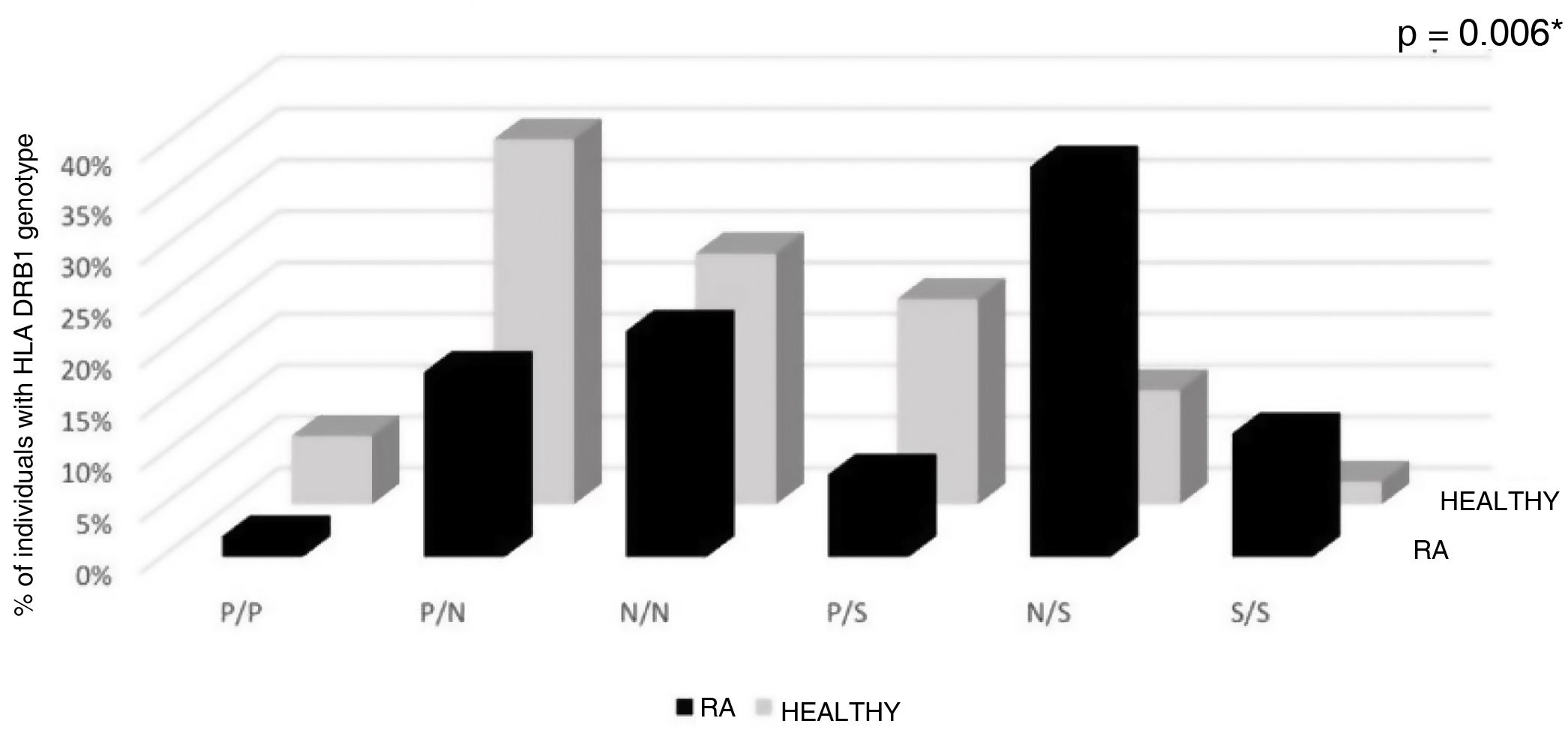
| de Vries genotype | 50 (100) | 100 | 45 (100) | 90 | *0.006 |
| P/P | 1 (2.00) | 2 | 3 (6.66) | 6 | |
| P/N | 9 (18.00) | 9(P)/9(N) | 16 (35.55) | 16(P)/16(N) | |
| N/N | 11 (22.00) | 22 | 11 (24.44) | 22 | |
| P/S | 4 (8.00) | 4(P)/4(S) | 9 (20.02) | 9(P)/9(S) | |
| N/S | 19 (38.00) | 19(N)/19(S) | 5 (11.11) | 5(N)/5(S) | |
| S/S | 6 (12.00) | 12 | 1 (2.22) | 2 | |
| Mattey genotype | 50 (100) | 100 | 45 (100) | 90 | 0.62 |
| D70EC−/D70SE− | 7 (14.00) | 14 | 12 (26.70) | 24 | |
| Q70EC−/D70SE− | 10 (20.00) | 10(Q70SE)/10(D70SE−) | 9 (20.00) | 9 (Q70SE−)/9(D70SE−) | |
| Q70EC−/Q70SE− | 4 (8.00) | 8 | 6 (13.33) | 12 | |
| Q70EC+/D70SE− | 9 (18.00) | 9(Q70SE+)/9(D70SE−) | 6 (13.33) | 6(Q70SE+)/6(D70SE−) | |
| Q70EC+/Q70SE− | 11 (22.00) | 11(Q70SE+)/11(Q70SE−) | 7 (15.55) | 7(Q70SE+)/7(Q70SE−) | |
| Q70EC+/Q70SE+ | 3 (6.00) | 6 | 1 (2.22) | 2 | |
| Other | 6 (12.00) | 12 | 4 (8.88) | 8 | |
| Tezenas du Montcel genotype | 47 (100) | 94 | 47 (100) | 94 | 0.19 |
| L/L | 21 (44.68) | 42 | 31 (65.95) | 62 | |
| S3P/L | 20 (42.55) | 20(S3P)/20(L) | 12 (25.53) | 12(S3P)/12(L) | |
| S2/L | 2 (4.27) | 2(S2)/2(L) | 3 (6.38) | 3(S2)/3(L) | |
| S3P/S3P | 3 (6.38) | 6 | 1 (2.12) | 2 | |
| S2/S2 | 0 | 0 | 0 | 0 | |
| S2/S3P | 1 (2.12) | 1(S2)/1(S3P) | 0 | 0 | |
| Raychaudhuri risk classification | RA 97 alleles | Healthy 92 alleles | 0.18 | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| (1) SSEA | 3 | 3.09 | 7 | 7.62 | |
| (2) SSKR | 3 | 3.09 | 7 | 7.62 | |
| (3) SGRL | 11 | 11.34 | 7 | 7.62 | |
| (4) LFEA | 2 | 2.06 | 3 | 3.10 | |
| (5) SSRE | 1 | 1.03 | 1 | 1.10 | |
| (6) SGRA/SSRA | 19 | 19.58 | 11 | 11.95 | |
| (7) GYRQ | 2 | 2.06 | 8 | 8.70 | |
| (8) PRAA | 6 | 6.18 | 8 | 8.70 | |
| (9) SSKA | 1 | 1.03 | 0 | 0.00 | |
| (10) VHEA | 1 | 1.03 | 1 | 1.08 | |
| (11) DFRE | 2 | 2.06 | 2 | 2.19 | |
| (12) VHRE | 18 | 18.57 | 17 | 18.50 | |
| (13) PRRA | 7 | 7.22 | 6 | 6.56 | |
| (14) LFRA | 4 | 4.14 | 7 | 7.62 | |
| (15) VFRA/VHRA | 15 | 15.46 | 5 | 5.45 | |
| (16) VHKA | 2 | 2.06 | 2 | 2.19 | |
RA: rheumatoid arthritis.
*p < 0.05 significant by Fisher’s exact and Chi-square tests.
With reference to the classic SE classification (Gregersen et al.), in the group with RA the highest percentage (46%) had SE+/SE− genotype, followed by 42% of negatives for SE, while 12% had SE+/SE+ genotype. In total, in the RA population, 58% had at least one allele positive for SE (allelic frequency of SE+ in RA = 35). In contrast, the majority of the healthy population was negative for SE (68.1%); 29.8% had SE+/SE− genotype, being SE+/SE+ a healthy subject; 32% of the healthy population had at least one allele positive for SE (allelic frequency of SE+ in healthy people = 16); these differences were statistically significant (p = 0.006) (Table 3, Fig. 1).
The classification of Vries et al. according to the genotypes S/S, N/S, P/S, N/N, P/N and P/P evidenced differences between the RA and the healthy population: 12% of the RA population had two susceptibility alleles, compared with 2% in the healthy population. 6.66% of the healthy population had two protective alleles, while in the RA population only 2% had these alleles The N/S genotype had a higher frequency in the population with RA, in turn, the P/N genotype was more frequent in the population without RA; the frequency of the N/N genotype was similar in both population groups. These findings were statistically significant (p = 0.006) (Table 3, Fig. 2).
Distribution of HLA DRB1 genotype in RA and healthy individuals according to the classification of de Vries et al. Aminoacids in positions HLA DRβ1 67, 70, 71, 73 and 74.
Genotypes P/P: protective/protective; P/N: protective/neutral; N/N: neutral/neutral; P/S: protective/susceptibility; N/S: neutral/susceptibility: S/S susceptibility/susceptibility.
*p < 0.05 significant by Fisher’s exact and Chi-square tests.
For the typification of the alleles, ambiguities were not found in 188 alleles (97 RA and 91 healthy) (Table 4). The most prevalent alleles in the RA group with respect to the healthy population were 14:02, 04:04, 08:02, 04:05 and 10:01. High frequencies of the alleles 07:01, 03:01, 13:02, 01:02 and 12:01 were found in healthy population, in comparison to the RA group. The alleles that accompanied the genotype in healthy individuals carriers of the allele 01:02 (five individuals) were SE− (classification of Gregersen et al.), (01:02/04:07, 01:02/03:01, 01:02/16:02, 01:02/13:02, 01:02/15:01). 50% of the individuals who carried the allele 08:02 had the allele 14:02 (08:02/15:01, 08:02/04:07, 08:02/14:02, 08:02/14:02, 08:02/08:02, 08:02/14:02) in their genotype.
Frequency of HLA DRB1 alleles in Colombian healthy individuals and with RA.
| Allele | RA | Healthy |
|---|---|---|
| HLA DRB1 alleles more frequent in individuals with RA | ||
| 14:02 | 10 | 2 |
| 04:04 | 8 | 2 |
| 08:02 | 7 | 4 |
| 04:05 | 4 | 1 |
| 10:01 | 4 | 1 |
| 04:03 | 3 | 1 |
| 16:01 | 3 | 1 |
| 04:11 | 2 | 0 |
| 11:02 | 1 | 0 |
| 12:07 | 1 | 0 |
| 13:03 | 1 | 0 |
| HLA DRB1 alleles more frequent in healthy individuals | ||
| 07:01 | 2 | 8 |
| 03:01 | 3 | 6 |
| 13:02 | 0 | 5 |
| 01:02 | 1 | 5 |
| 12:01 | 1 | 4 |
| 03:02 | 0 | 1 |
| 15:03 | 0 | 1 |
| HLA DRB1 alleles with similar frequency in individuals with RA and healthy | ||
| 04:07 | 12 | 16 |
| 15:01 | 5 | 5 |
| 11:01 | 5 | 3 |
| 16:02 | 4 | 5 |
| 01:01 | 3 | 2 |
| 01:03 | 2 | 3 |
| 04:01 | 2 | 2 |
| 08:04 | 2 | 2 |
| 09:01 | 2 | 2 |
| 11:04 | 2 | 2 |
| 13:01 | 2 | 2 |
| 08:01 | 2 | 1 |
| 15:02 | 1 | 2 |
| 04:02 | 1 | 1 |
| 14:01 | 1 | 1 |
RA, rheumatoid arthritis.
The 04:07, 15:01, 11:01, 16:02 and 01:01 alleles had similar representation in RA and healthy populations (Table 4).
DiscussionThis is the first study that compares different classification methods for HLA DRB1 alleles related to RA in the Colombian population. Many analyzes have focused on the region of amino acids referred to as SE and its influence on the pathogenesis of RA, including susceptibility, severity, and response to treatment. Several modifications to the initial proposal, restricted to amino acids 70–74 of the third hypervariable region of HLA DRB1, have been proposed.19 Of the five systems of classification of HLA DRB1 used in this study, those of Gregersen et al. and de Vries et al. were adequate for the RA characterization in this population. The classic classification of SE (Gregersen et al.)6 was effective, simple and significant in the differentiation between RA and healthy people. The alleles related with SE according to this system have been a historical referent for an adequate determination of risk for RA related to HLA DR in the majority of studies and for diverse ethnic groups, and have been used before the emerging proposals of the new classification systems.8–10,20,21
The research by Nick de Vries et al.5 was conducted in a cohort of Dutch Caucasian subjects, and was adequate for the allocation of risk for RA in the population of this study; our findings regarding the system of de Vries et al. are the first reported in mestizo population. This system was applied by Morgan et al.16 in Caucasians from the United Kingdom, and it was determined a similar protective effect of the N and P alleles, which were considered part of the same group. Likewise, in the analysis of the genotypes, the proposed hypothesis of protection against RA by P alleles was denied.
Similarly to the findings of Tezenas du Montcel et al.,15 our results did not demonstrate a relationship of the positions 72–74 with risk for RA; however, we did not find a relationship of the disease with the risk genotypes. The method of Tezenas du Montcel et al. has been adequate in other Caucasian populations according to what has been found by Morgan et al.16 when associating S2 and S3P alleles with RA in residents in the United Kingdom, but not in this group of Colombian population.
The method proposed by Mattey et al.7 involved population from Spain and the United Kingdom. In our sample, the classification of Mattey was not adequate in the characterization of RA population. According to the results of this research, it was also not possible to confirm the dominant protective effect of the aminoacid D70 associated with SE−, reported by the authors when it was incorporated to the Q70SE+/D70SE genotype. We found a higher frequency of the DR4 haplotype in individuals with RA, similarly to the findings of the authors,7 who reported this higher relationship for the United Kingdom with respect to Spain; however, among the Spanish people there was a high frequency of Q70SE+DR4. Morgan et al.16 found adequate the Mattey classification system when it was applied in Caucasians residing in the United Kingdom.
The classification by Raychaudhuri et al. was not adequate in the differentiation of RA/healthy populations in our population, which could be due to the fact that there are 16 groups of haplotypes for the definition of risk for RA, while in this study there was a limited population size. Likewise, the HLA-DRB1*14:02 allele, positive for SE+ was related with RA in this and other research in various population groups6,7,15; however, Raychaudhuri et al. considered it of low risk.
Another important point to analyze is the frequency and significance of the HLA DRB1 alleles in RA and healthy populations. Among the most prevalent alleles in the RA group were HLA-DRB1*14:02, 04:04, 04:05 and 10:01, all carriers of the SE.22 Likewise, Spinel et al.8 found the HLA-DRB1*14:02 allele two times more frequent in RA than in controls in Colombian population; the HLA-DRB1*04:04 allele has been characteristic in Caucasians, associated with risk for the disease.9,23 It has been reported risk for RA with the presence of the HLA-DRB1*10:01 allele in various populations,5,14,15 which was also found with high frequency in Colombian RA population, according to the results of this study. In contrast, Morgan et al.16 in their study with Caucasian population found no association between the HLA-DRB1*10:01 allele and the disease.16 According to Mattey et al., this allele is classified as intermediate risk.7
Gilbert et al.24 related the HLA-DRB1*08:02 allele with a protective effect (but not significant) for RA, while de Vries et al.5 considered it to have a neutral effect. Although in the current research the DRB1*08:02 allele had a higher frequency in RA patients than in healthy individuals, 50% of the patients who had this allele and RA carried the DRB1*14:02 allele in their genotype, which could influence on a greater representation in population with RA, being the DRB1*08:02 allele characteristic of Colombian Amerindian populations.25
High frequencies of the alleles HLA-DRB1*07:01, 03:01, 01:02 and 13:02 were found in the healthy population, compared with the RA group. The alleles HLA-DRB1*07:01, 03:01 and 13:02 are SE−, unlike the allele 01:02, which is SE+. The protective effect of the HLA-DRB1*07:01 allele was confirmed by de Vries et al.5 and Raychaudhuri et al.,14 while the HLA-DRB1*01:02 allele, as well as the HLA-DRB1*01:01 allele, have been related to susceptibility to RA in various population groups, they code for SE and have been associated with part of the most important genetic risk in the development of the disease.6,7,15 In this population, in particular, the HLA-DRB1*01:02 allele was more representative in healthy population, while the HLA-DRB1*01:01 allele, characteristic of risk in different populations, presented a similar frequency in RA and in healthy individuals. The healthy population carrier of the HLA-DRB1*01:02 allele, however, did not present another susceptibility allele in their genotype.
One of the healthy individuals carried the HLA-DRB1*01:02/13:02 genotype; being the HLA-DRB1*13:02 allele exclusive of the healthy population in this study. The protective effect of the HLA-DRB1*13:02 allele has been confirmed in several studies, such as that conducted by Gibert et al.,24 who associated the HLA-DRB1*13:02 allele with protection against RA in French population, due to the negative charge in the P4 pocket; the authors reported this effect despite the relationship with risk alleles within the genotype. Another healthy individual who was positive for the HLA-DRB1*01:02 allele presented in his genotype the HLA-DRB1*15:01 allele. In the analysis of the relationship of the HLA-DRB1*15:01 allele, De Vries et al.5 associated this allele with significant protective effects, attributed to the presence of isoleucine in the position 67 HLA DRβ1. In addition, another genotype present in healthy population with the HLA-DRB1*01:02 allele was accompanied by the HLA-DRB1*03:01 allele; among the non RA individuals of this study there was a high prevalence of this last allele, which has commonly been considered as protective.7,14,15 Conversely, de Vries et al. associated the HLA-DRB1*03:01 allele with risk for RA in Caucasian population.5
The HLA-DRB1*04:07, 15:01 and 16:02 alleles had similar frequencies in healthy and RA populations. The HLA-DRB1*04:07 allele (SE−) had a “neutral effect” according to de Vries et al.5; and it was also classified by the model of Revirón et al., due to the positive electric charge in the P4 pocket,26 which coincides with the findings suggested in the present study.5 Raychaudhuri et al.,14 for their part, classified it into the risk category. The HLA-DRB1*16:02 allele is negative for SE according to the classic classification of Gregersen,6 while it is classified as neutral according to de Vries et al.5 and the model of Revirón et al.,26 similarly to our research, and it is categorized as low risk by the other authors.7,15 With respect to the HLA-DRB1*15:01 allele, we observed the same frequency in the healthy and RA populations, with a tendency to be a neutral allele, a classification also assigned by Raychaudhuri et al. (OR = 1); however, de Vries et al.5 reported it as a protective allele.
Regarding the presence of anti-CCP, it was exclusive to the population with RA in this research; positive anti-CCP tests predict the development of RA, often years before clinical confirmation of the disease.18 There is evidence of the high affinity in the interaction between citrullinated peptides and the molecules that share the SE.22 A trend towards the presence of at least one SE+ allele in the production of anti-CCP (63% of the subject anti-CCP with SE+) was observed in this study; nevertheless; there was a representative percentage of RA individuals anti-CCP+ who were negative for SE, similarly to the findings of Balsa et al.9 in their study in Spanish population.
The possibility of generating anti-CCP by alleles other than SE may be due in part to the fact that even when the SE+ alleles are related with HLA DRβ1 proteins with highly positive charge in the P4 pocket, the citrullinated peptides may also bind to other sequences of peptide anchor, beyond those that carry the sequence of the SE.27 In addition, other genes apart from the MHC have been associated with citrulline-induced autoimmunity. Morgan et al.28 found the PTPN22 gene fundamentally associated with anti-CCP; in the same way, the presentation of citrullinated peptides has been identified with HLA DQ alleles.29,30
Among the strengths of the current research, our findings regarding classification methods for SE are the first reported in the mestizo population and, additionally, information related to the most frequent alleles in Colombian individuals with RA is provided. We consider that the weaknesses associated with the research were mainly due to the reduction of more than 30% in the sample size, due to the fact that the current or past smoking habit was taken into account, being a variable that must be controlled in the model of the RA and the SE.
ConclusionsOur findings demonstrate that while alternative SE classifications appear to be useful in the populations in which they have been initially tested, they are not fully applicable to all population groups; there is a methodological diversity among the studies for the determination of the HLA DRB1 alleles, which also represent a highly polymorphic system. The systems of Gregersen et al. and de Vries et al. were adequate for the characterization of RA in this group of Colombian population; our results have clinical relevance, taking into account the genetic differences of our population in relation to the findings in other latitudes. Likewise, this research provides information related to the most frequent alleles in Colombian individuals who present this pathology; the foregoing, in the context of multiple risk factors, not only environmental but also genetic. Finally, we consider that our results could be complementary clinical support tools, useful to stratify the individual risk in our population and in individuals with a family history of RA; nevertheless, we consider pertinent the reproduction of the current study in a larger population group; as well as in other mestizo populations in which they have not yet been tested.
FundingThis study was supported by the Division of Research from Bogotá, Universidad Nacional de Colombia (Hermes Code: 20166), the Administrative Department of Science, Technology and Innovation Colciencias (N.o 130854531734-2011), Hospital Militar Central (2014–4337), Universidad El Bosque (UB-293–2011), Bogotá, Colombia.
Conflict of interestThe authors declare that they have no conflict of interest.
We thank Dr. Víctor Hidalgo for the statistical analyses, Dr. Wilson Bautista for his contributions and guidance in writing from the area of rheumatology, Dr. Mauricio Rodríguez for the methodological approach, the Universidad el Bosque and Dr. Rafael Valle for facilitating the spaces for the development of the project in the Service of Rheumatology of the Hospital Militar Central.