Identification of dermatophytes is usually performed through morphological analyses. However, it may be hindered due to the discovery of new species and complexes and, with some isolates, by the absence of fructification. Matrix-assisted laser desorption/ionisation-time-of-flight mass spectrometry (MALDI-TOF MS) seems to be an option for improving identification.
AimsTo develop a database (DB) for the identification of dermatophytes with MALDI-TOF MS, including 32 isolates from the Red de Micología de la Ciudad Autónoma de Buenos Aires [Mycology Network of the Autonomous City of Buenos Aires] (RMCABA) and one reference isolate (RMCABA DB), and evaluate its performance when added to the DB from the supplier, Bruker (Bruker DB).
MethodsAll the isolates in the RMCABA DB were identified based on morphology and sequencing. To evaluate the performance of the extended DB (Bruker DB plus RMCABA DB), 136 clinical isolates were included.
ResultsThe percentages of identification at the species level increased from 45% to 88%, but the identification at the genus level decreased from 23% to 7%.
ConclusionsMALDI-TOF MS yielded better performance in the identification of dermatophytes after including the RMCABA DB, which encompassed local isolates.
La identificación de dermatofitos se realiza, usualmente, a través del análisis morfológico, pero puede verse dificultada por la aparición de nuevas especies y complejos de especies y, en algunos aislamientos, por la falta de elementos de fructificación. Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) aparece como una alternativa para mejorar la identificación.
ObjetivosConstruir una base de datos (BaDa) para la identificación de dermatofitos mediante MALDI-TOF MS, con 32 aislamientos de la Red de Micología de la Ciudad Autónoma de Buenos Aires (RMCABA) y una cepa de referencia (BaDa RMCABA), y evaluar su desempeño al incorporarla a la BaDa del proveedor Bruker (BaDa Bruker).
MétodosTodos los aislamientos incorporados a la BaDa RMCABA se identificaron por morfología y secuenciación. Para la evaluación de la BaDa ampliada (BaDa Bruker más BaDa RMCABA) se incluyeron 136 aislamientos clínicos.
ResultadosEl porcentaje de identificación a nivel de especie se incrementó del 45 al 88%, mientras que la identificación a nivel de género disminuyó del 23 al 7%.
ConclusionesCon la incorporación de la BaDa RMCABA, que contó con aislamientos regionales, se observó un mejor desempeño en la identificación de dermatofitos por MALDI-TOF MS.
Infections caused by dermatophytes are highly prevalent throughout the world. Direct examination enables a rapid diagnosis of dermatophytosis and, thus, the immediate prescription of a treatment, while culture has an epidemiological value and confirms the diagnosis with certainty. The fungal species is usually determined through its macro- and micromorphological characteristics. This procedure depends on the experience of the technician and the development of the fungus in the cultures. Identification may be hampered by the description of new species and species complexes, many of which are phenotypically similar but different in terms of pathogenesis, since they may have different proteolytic capacity depending on the reservoir.1,6,8,9
New methodologies such as genomics and proteomics have been incorporated in laboratories, emerging as diagnostic tools capable of quickly and reliably identifying microorganisms. At present, the matrix-assisted laser desorption/ionisation-time-of-flight mass spectrometry (MS) technique has been quickly introduced in numerous clinical microbiology laboratories as a new technological resource. Its usefulness in the identification of bacteria and yeasts has been proven; however, it still has some limitations when identifying mycelial fungi.2,10,16,23,24 The heterogeneity of the results obtained with filamentous fungi is likely linked to the different protein profiles, depending on the different characteristics of the culture, the media used and the incubation time.3 The results are improved with the implementation of protocols in which these variables are standardised.14 Different publications have shown that, in order to identify mycelial fungi efficiently, it is important to have a comprehensive, complete database that includes a large number of protein profiles for each species.10,11,16,18,21 The library from the manufacturer Bruker (MBT DB 5627 MSP list, Filamentous Fungi Library 1.0, Bruker Daltonics) has a small number of dermatophyte spectra. Therefore, the creation of a database (DB) with regional isolates would enable an improvement in the identification.
The objectives of the study were the following: to develop a DB for the identification of dermatophytes by means of MALDI-TOF MS, using regional isolates from Red de Micología de la Ciudad Autónoma de Buenos Aires (Mycology Network of the Autonomous City of Buenos Aires [RMCABA] – RMCABA DB) and an external control strain; to evaluate an extended DB (Bruker DB plus RMCABA DB); and to compare its performance to that of the DB provided by the manufacturer (Bruker DB).
Materials and methodsCreation of the RMCABA DBThe fungi were identified by their macro- and micromorphological (MA) phenotypic characteristics, following the keys of the Atlas of Clinical Fungi by de Hoog et al.7 and by sequencing the intergenic spacers of ribosomal DNA using primers ITS1 and ITS4.4,12,15 Thirty-three (33) dermatophytes, with the name according to the Hoog et al.’s taxonomic update in parentheses, were included6: Epidermophyton floccosum, n=2; Microsporum canis, n=3; Microsporum gypseum (Nannizzia gypsea), n=1; Microsporum nanum (Nannizzia nana), n=1; Microsporum fulvum (Nannizzia fulva), n=1; Trichophyton mentagrophytes complex, n=10 (this study includes the species Trichophyton interdigitale and Trichophyton mentagrophytes); Trichophyton rubrum, n=5; Trichophyton tonsurans, n=5; Trichophyton violaceum, n=1; Trichophyton schoenleinii, n=1; Trichophyton verrucosum, n=1; Trichophyton ajelloi (Arthroderma uncinatum), n=1; and Trichophyton erinacei (DMic 175540 UK NEQAS; international quality expertise), n=1. The isolates were subcultured on Sabouraud glucose agar with chloramphenicol (Biokar Diagnostics, France) and incubated at 28°C for 5–7 days. Growth was considered optimal when the colony had a diameter of 5mm.
Protein extractionProtein extraction both for the set up of the DB and the analysis of the extended DB was performed based on the methodology published by Lau et al.13 A portion of the colony was put in an Eppendorf tube containing 250μl of absolute ethanol (Sigma-Aldrich, Lyon, France) and 50μl of zirconia/silica beads (0.1mm diameter). The tube was shaken for 15min and centrifuged at 13,000rpm (2min) and the supernatant was discarded. Fifty (50) μl of 70% formic acid (Sigma-Aldrich, Lyon, France) were added to the pellet and stirred for 5min, then 50μl of 100% acetonitrile (Sigma-Aldrich, Lyon, France) were added. This mixture was stirred for 5min and centrifuged at 13,000rpm (2min); 1μl of the supernatant was placed on the metal plate and covered with 1μl of alpha-cyano-4 hydroxy-cinnamic acid matrix (Bruker Daltonics, Germany).
The development of the main protein profiles (main spectra profile [MSP]) was done according to the instructions in the Bruker guide (Custom MSP and library creation, EE, 2/2014). For the MALDI-TOF MS analysis, Bruker Microflex LT, Maldi Biotyper software, version 3.1, was used. Identification was evaluated with the Bruker DB and then with the extended DB.
Evaluation of extended DBOne hundred and thirty-six (136) isolates from RMCABA hospitals were included, all previously identified by MA: E. floccosum, n=1; M. canis, n=11; M. gypseum (N. gypsea), n=7; M. nanum (N. nana), n=1; M. fulvum (N. fulva), n=2; T. mentagrophytes complex, n=25; T. rubrum, n=74; T. tonsurans, n=14; and T. verrucosum, n=1. The scores used as identification criteria were: ≥1.7, identification at the species level, 1.50–1.69, identification at the genus level, <1.50, no identification (no ID). Discrepancy (discrepant ID) was considered when MALDI-TOF MS yielded an identification different from that obtained with MA; in this situation sequencing was carried out.
The data were processed with R, version 4.0.2. The kappa statistic was calculated with the Psych package. The Wilcoxon test was used to compare the means of the scores. The receiver operating characteristic (ROC) curve analysis was performed by varying the cut-off point based on the score obtained. Sensitivity and specificity were calculated for the cut-off point ≥1.5 (R Foundation for Statistical Computing, Vienna, Austria; https://www.R-project.org).
ResultsThe identification with the Bruker DB of the dermatophytes selected to make up the RMCABA DB showed a percentage of ID at the species level of 36% (12/33), a percentage of ID at the genus level of 21% (7/33), a percentage of no ID of 27% (9/33) and a percentage of misidentification of 15% (5/33), which corresponded to the T. mentagrophytes complex (Table 1). The Cohen kappa statistic for estimating the agreement between the Bruker DB and the MA identification was estimated at 0.65 (confidence interval [CI] 95%, 0.56–0.74).
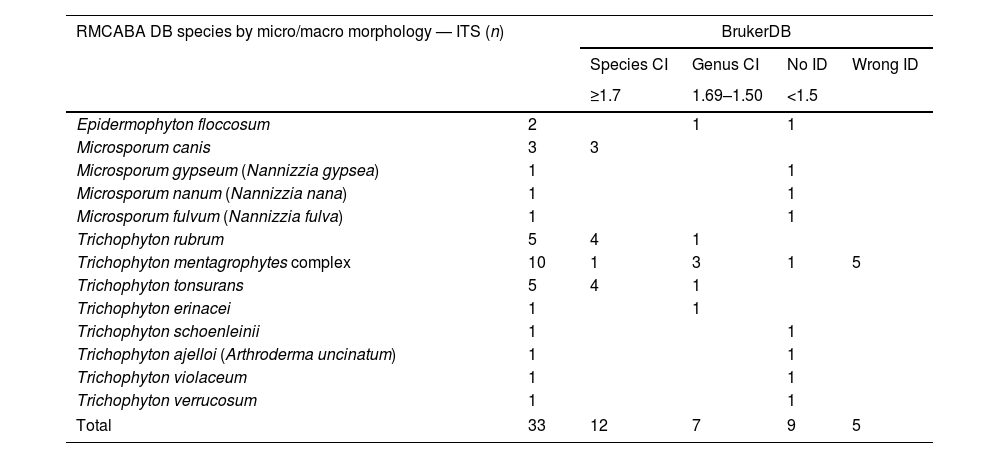
Analysis of the dermatophytes in the RMCABA DB with the database from the supplier Bruker.
| RMCABA DB species by micro/macro morphology — ITS (n) | BrukerDB | ||||
|---|---|---|---|---|---|
| Species CI | Genus CI | No ID | Wrong ID | ||
| ≥1.7 | 1.69–1.50 | <1.5 | |||
| Epidermophyton floccosum | 2 | 1 | 1 | ||
| Microsporum canis | 3 | 3 | |||
| Microsporum gypseum (Nannizzia gypsea) | 1 | 1 | |||
| Microsporum nanum (Nannizzia nana) | 1 | 1 | |||
| Microsporum fulvum (Nannizzia fulva) | 1 | 1 | |||
| Trichophyton rubrum | 5 | 4 | 1 | ||
| Trichophyton mentagrophytes complex | 10 | 1 | 3 | 1 | 5 |
| Trichophyton tonsurans | 5 | 4 | 1 | ||
| Trichophyton erinacei | 1 | 1 | |||
| Trichophyton schoenleinii | 1 | 1 | |||
| Trichophyton ajelloi (Arthroderma uncinatum) | 1 | 1 | |||
| Trichophyton violaceum | 1 | 1 | |||
| Trichophyton verrucosum | 1 | 1 | |||
| Total | 33 | 12 | 7 | 9 | 5 |
RMCABA DB: database from the Red de Micología de la Ciudad Autónoma de Buenos Aires [Mycology Network of the Autonomous City of Buenos Aires]; Bruker DB: database from the supplier Bruker; correct identification on the species level (species CI); correct identification on the genus level (genus CI); no identification (no ID); misidentification (wrong ID).
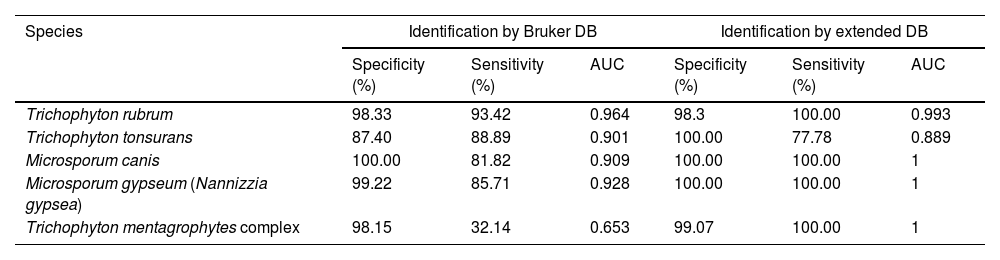
The intensities and position (m/z) of the MSPs for each genus and species included in the RMCABA DB were compared in the different species of dermatophytes (Fig. 1a and b). High intraspecies similarity and differences between species were observed, except for T. mentagrophytes and T. interdigitale, which were considered part of the T. mentagrophytes complex, as other authors have also mentioned.6,24
Intensities and position (m/z) of the main peaks in the spectra of some species. (a) Epidermophyton floccosum (blue), Microsporum canis (red), Microsporum fulvum (Nannizzia fulva) (purple), Microsporum gypseum (Nannizzia gypsea) (green), Microsporum nanum (Nannizzia nana) (light blue). (b) Trichophyton tonsurans (blue), Trichophyton rubrum (red), Trichophyton mentagrophytes complex (light blue), Trichophyton violaceum (green), Trichophyton schoenleinii (yellow), Trichophyton erinacei (purple), Trichophyton verrucosum (orange) and Trichophyton ajelloi (Arthroderma uncinatum) (brown).
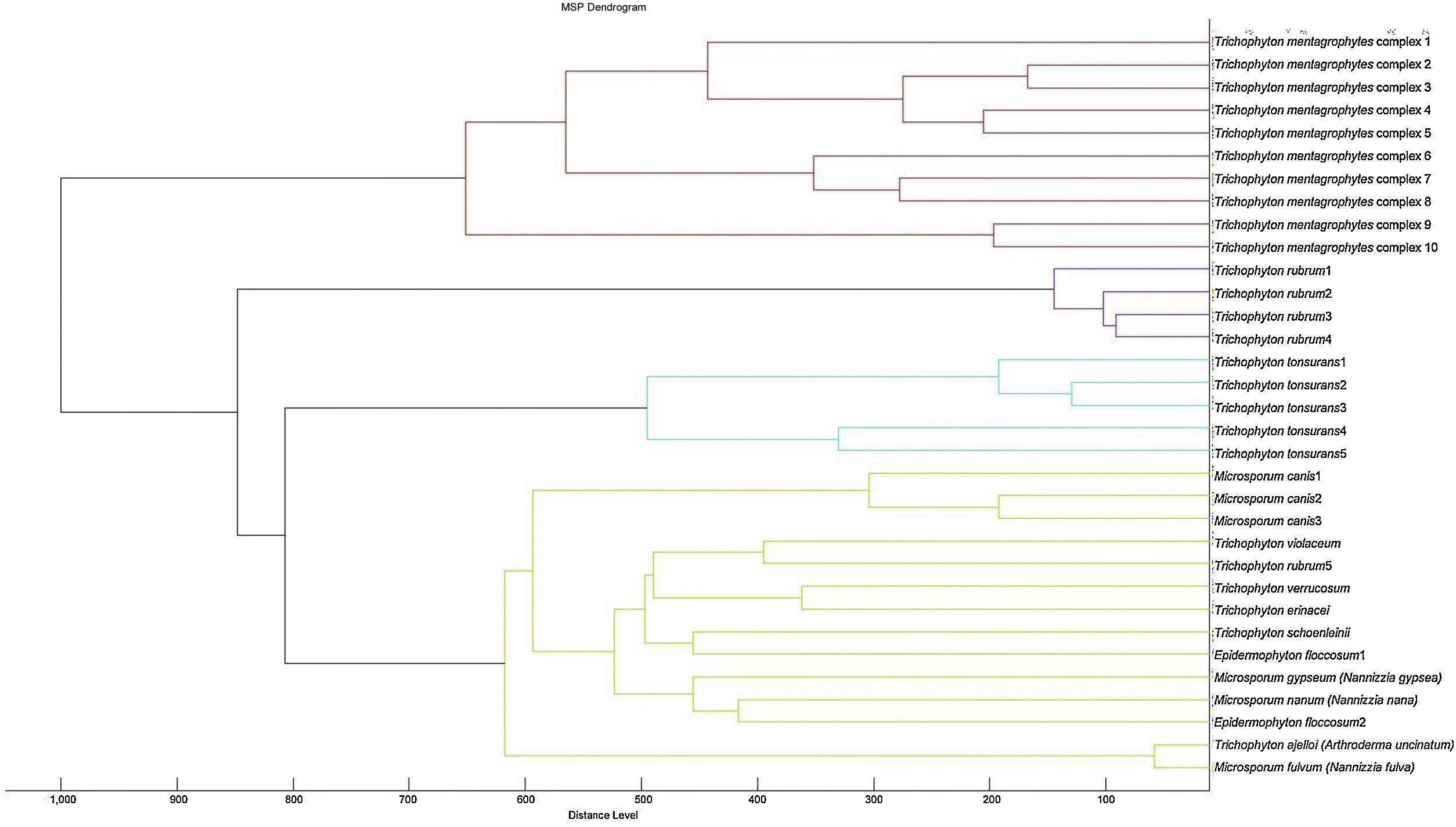
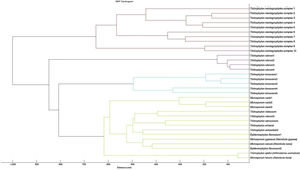
A cluster analysis of the spectra obtained from the isolates used for the RMCABA DB was performed (Fig. 2). Zero indicates complete similarity, and 1000 indicates complete dissimilarity. When an arbitrarily chosen distance level of 700 is applied, the T. mentagrophytes, T. rubrum, and T. tonsurans isolates cluster in different groups, while no groups could be observed among the remaining species due to the small number of isolates.
Table 2 shows the distribution of the scores obtained with the Bruker DB and the extended DB. With the Bruker DB, a percentage of ID at the species level of 45% (61/136), a percentage of ID at the genus level of 23% (31/136), a percentage of no ID of 24% (32/136) and a percentage of discrepant ID of 9% (12/136) were obtained. When the RMCABA DB was added, the percentages were as follows: ID at the species level was 88% (120/136), ID at the genus level was 7% (10/136), no ID was 0% and discrepant ID was 4% (6/136); a statistically significant increase in scores was observed (Wilcoxon [W]=15703; p<0.001). Eighty-five per cent (85%) (115/136) of the isolates were first identified with RMCABA DB MSP. The Cohen kappa statistic for estimating the agreement between the Bruker DB and the MA identification was 0.98 (95% CI, 0.94–1.0). The agreement between the Bruker DB analysis and the extended DB was 0.65 (95% CI, 0.56–0.74) (Table 3) (Fig. 3a and b).
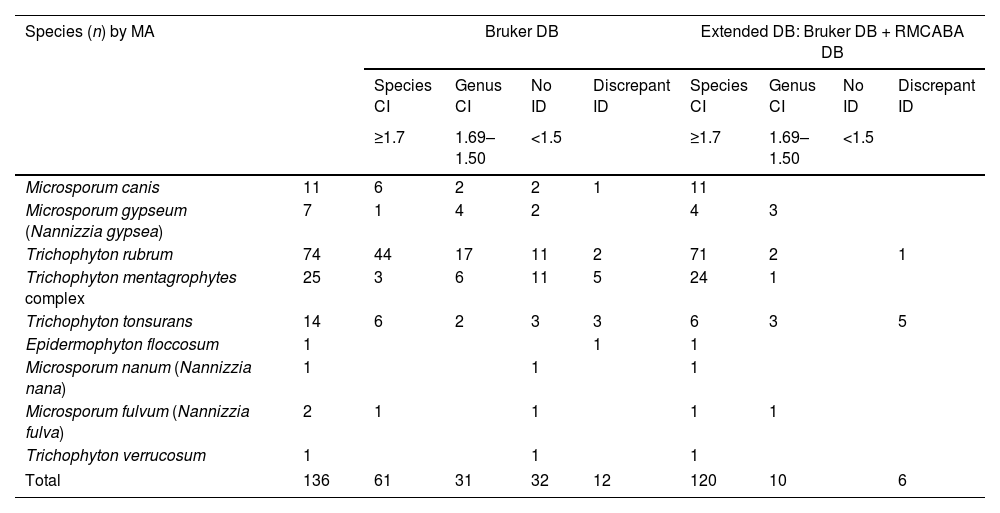
Scores obtained with the Bruker DB and the extended DB.
| Species (n) by MA | Bruker DB | Extended DB: Bruker DB + RMCABA DB | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Species CI | Genus CI | No ID | Discrepant ID | Species CI | Genus CI | No ID | Discrepant ID | ||
| ≥1.7 | 1.69–1.50 | <1.5 | ≥1.7 | 1.69–1.50 | <1.5 | ||||
| Microsporum canis | 11 | 6 | 2 | 2 | 1 | 11 | |||
| Microsporum gypseum (Nannizzia gypsea) | 7 | 1 | 4 | 2 | 4 | 3 | |||
| Trichophyton rubrum | 74 | 44 | 17 | 11 | 2 | 71 | 2 | 1 | |
| Trichophyton mentagrophytes complex | 25 | 3 | 6 | 11 | 5 | 24 | 1 | ||
| Trichophyton tonsurans | 14 | 6 | 2 | 3 | 3 | 6 | 3 | 5 | |
| Epidermophyton floccosum | 1 | 1 | 1 | ||||||
| Microsporum nanum (Nannizzia nana) | 1 | 1 | 1 | ||||||
| Microsporum fulvum (Nannizzia fulva) | 2 | 1 | 1 | 1 | 1 | ||||
| Trichophyton verrucosum | 1 | 1 | 1 | ||||||
| Total | 136 | 61 | 31 | 32 | 12 | 120 | 10 | 6 | |
RMCABA DB: database from the Red de Micología de la Ciudad Autónoma de Buenos Aires [Mycology Network of the Autonomous City of Buenos Aires]; Bruker DB: database from the supplier Bruker; correct identification at species level (species CI); correct identification at genus level (genus CI); no identification (no ID); discrepant identification (discrepant ID).
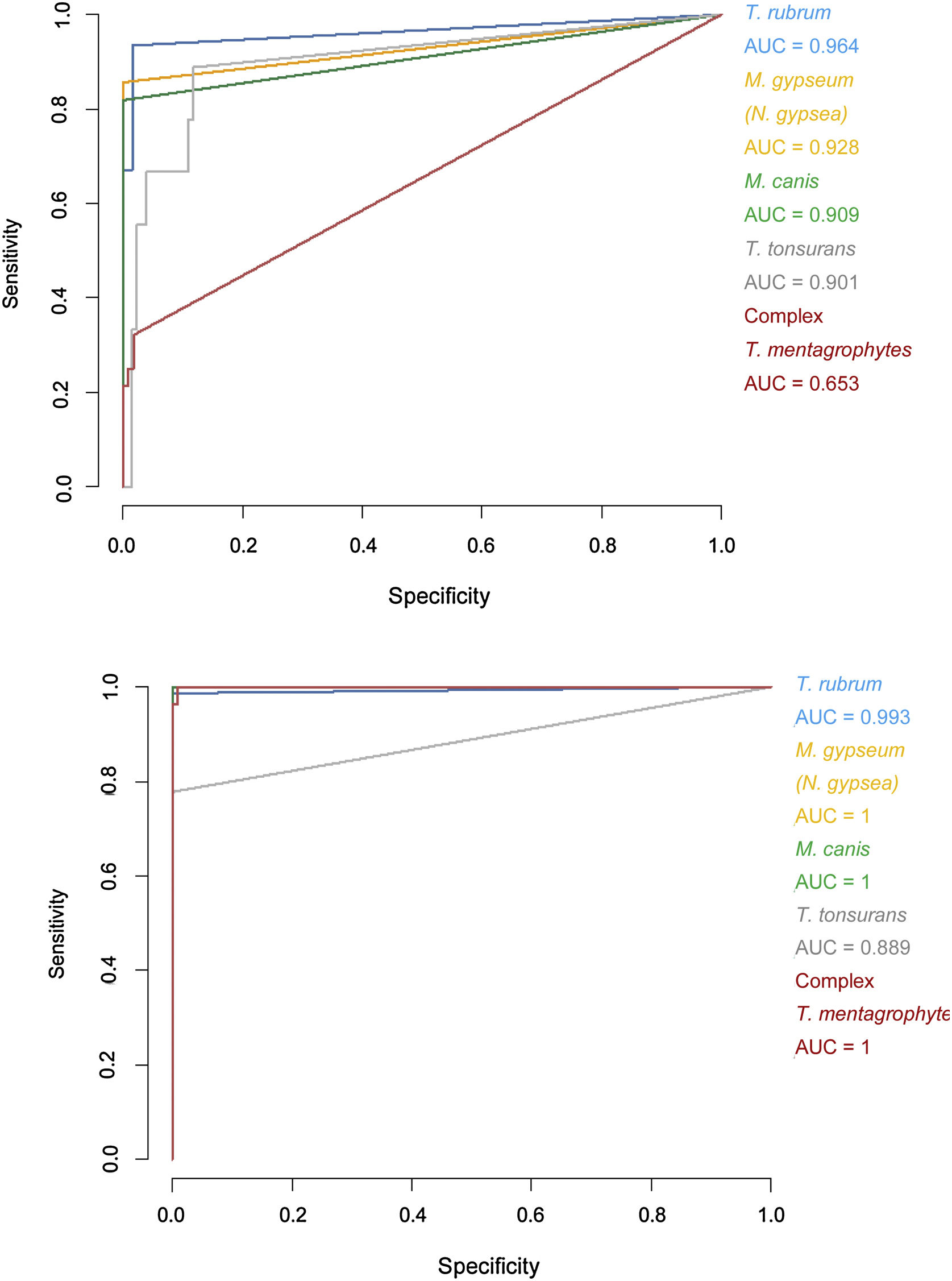
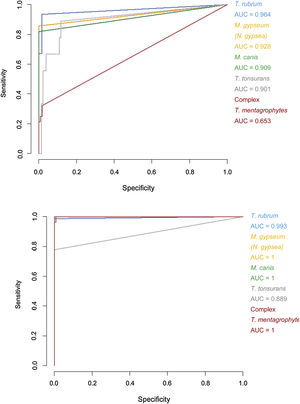
Specificity and sensitivity percentages considering the minimum scores (>1.5) and areas under the curve (AUC) for the receiver operating characteristic (ROC) curves of the Bruker DB versus those of the extended DB.
| Species | Identification by Bruker DB | Identification by extended DB | ||||
|---|---|---|---|---|---|---|
| Specificity (%) | Sensitivity (%) | AUC | Specificity (%) | Sensitivity (%) | AUC | |
| Trichophyton rubrum | 98.33 | 93.42 | 0.964 | 98.3 | 100.00 | 0.993 |
| Trichophyton tonsurans | 87.40 | 88.89 | 0.901 | 100.00 | 77.78 | 0.889 |
| Microsporum canis | 100.00 | 81.82 | 0.909 | 100.00 | 100.00 | 1 |
| Microsporum gypseum (Nannizzia gypsea) | 99.22 | 85.71 | 0.928 | 100.00 | 100.00 | 1 |
| Trichophyton mentagrophytes complex | 98.15 | 32.14 | 0.653 | 99.07 | 100.00 | 1 |
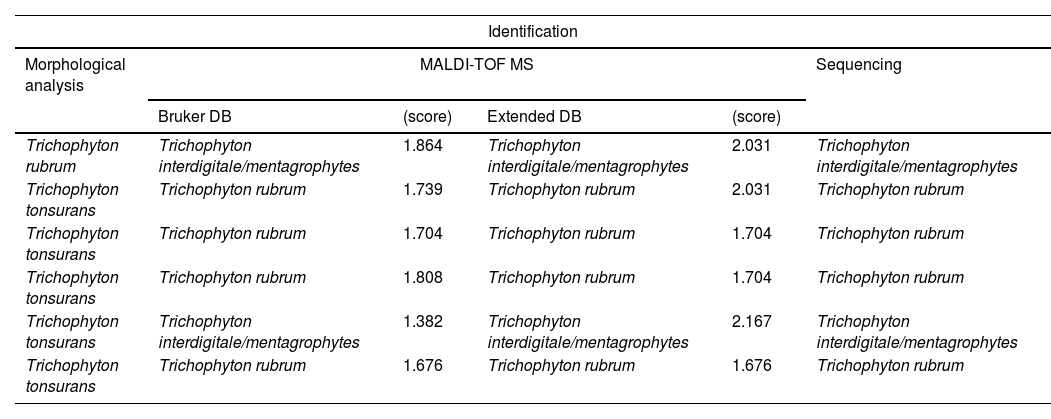
The increase in scores was particularly notable in the T. mentagrophytes complex, in which 24 out of 25 isolates were identified with the extended DB. Adding spectra of species not present in the Bruker DB enabled us to correctly identify some clinical isolates (Table 2). The discrepancies between the results obtained using the extended DB and the MA were analysed by sequencing, and resolved in favour of MALDI-TOF MS in all cases, corresponding to isolates formerly identified as T. tonsurans (5 isolates) and T. rubrum (1 isolate) (Table 4).
Analysis of the discrepancies according to the different methodologies.
| Identification | |||||
|---|---|---|---|---|---|
| Morphological analysis | MALDI-TOF MS | Sequencing | |||
| Bruker DB | (score) | Extended DB | (score) | ||
| Trichophyton rubrum | Trichophyton interdigitale/mentagrophytes | 1.864 | Trichophyton interdigitale/mentagrophytes | 2.031 | Trichophyton interdigitale/mentagrophytes |
| Trichophyton tonsurans | Trichophyton rubrum | 1.739 | Trichophyton rubrum | 2.031 | Trichophyton rubrum |
| Trichophyton tonsurans | Trichophyton rubrum | 1.704 | Trichophyton rubrum | 1.704 | Trichophyton rubrum |
| Trichophyton tonsurans | Trichophyton rubrum | 1.808 | Trichophyton rubrum | 1.704 | Trichophyton rubrum |
| Trichophyton tonsurans | Trichophyton interdigitale/mentagrophytes | 1.382 | Trichophyton interdigitale/mentagrophytes | 2.167 | Trichophyton interdigitale/mentagrophytes |
| Trichophyton tonsurans | Trichophyton rubrum | 1.676 | Trichophyton rubrum | 1.676 | Trichophyton rubrum |
The identification of microorganisms by means of MALDI-TOF MS is based on acquiring a protein profile (2–20kDa) and comparing it to a DB with reference spectra.22 The DB for the identification of mycelial fungi provided by Bruker is limited and contains only the most common species of dermatophytes. Hence, an expanded DB with mostly regional isolates was made. L’Ollivier et al. analysed the accuracy of MALDI-TOF MS in the identification of dermatophytes from 10 studies published between 2008 and 2015: the identification values ranged from 14% to 100%. The authors attributed this variability to differences in the protein extraction process and the DB used. All the studies showed that the use of an in-house DB improves the performance of MALDI-TOF MS for the identification of dermatophytes.17 Packeu et al. compared the Bruker system to a DB developed by them for the identification of 176 clinical isolates of dermatophytes, and achieved a 90% of correct identifications versus a 14% with the supplier's DB. In addition, if lower scores than those recommended by the manufacturer were considered, then these percentages increased to 97% for the in-house DB and 54% for the manufacturer's DB.22 Other authors such as Calderaro et al. determined the ability of an in-house DB with the Bruker system to identify 64 clinical isolates of dermatophytes; using a score >2, 73% (47/64) of the isolates were correctly identified, and this percentage raised to 100% if the score considered was 1.7–2. They also evaluated the incubation time and found that increasing the incubation time, and having carried out a previous extraction procedure, improved the percentage of correct identifications.2 Karabicak et al. showed that using an in-house DB and lower scores improved the identification of dermatophytes by MALDI-TOF MS.11 Da Cunha et al. evaluated their own DB with the Bruker system and the usefulness of the direct transfer method, and found the identification of dermatophytes with Bruker's DB to be lacking, while adding their DB yielded 97% agreement with sequencing in 276 isolates. The direct transfer method enabled the identification of 85% of the isolates.5,11,23
When we analysed with the Bruker DB the 33 isolates used in the RMCABA DB, poor values (36%) were obtained in the identification at the species level due to both the low scores and the misidentifications. On the other hand, some of the species, T. schoenleinii, T. verrucosum, T. ajelloi (A. uncinatum) and M. nanum (N. nana), were not present in the supplier's DB. With the implementation of an extended DB the percentages of identification improved at the species level and there were not isolates with no ID. It is important to note that 85% of the clinical isolates were properly identified and they matched with the first spectrum provided with the extended DB. The score that was considered suitable for the identification at the species level was ≥1.7. This value is lower than that used for bacteria and that recommended by the manufacturer, but it has already been used by several authors for both yeasts and mycelial fungi.2,11,22 As previously mentioned, many publications have shown that using lower scores and adding an in-house DB, improves the identification of dermatophytes by MALDI-TOF MS.20,21,23 In our study, a statistically significant increase in scores (p<0.05) was seen with the implementation of the RMCABA DB.
There were discrepancies in the identification of common species of dermatophytes, such as T. rubrum and T. mentagrophytes complex, between MA and MALDI-TOF MS (Table 4). The isolates involved were analysed by sequencing and the identification was resolved in favour of MALDI-TOF MS in all cases. This led us to consider that, even with these species, the morphological analysis may fail, especially with species complexes and new taxonomic rearrangements based on genomics and natural reservoirs of dermatophytes.
When comparing the intensities and the m/z position of the peaks of the different species with the RMCABA DB, the spectra showed intraspecies similarities and it was possible to distinguish between species with the exception of those within T. mentagrophytes complex, which were identified by ITS sequencing as T. interdigitale. Suh et al., in a study of dermatophytes of this complex identified by means of MALDI-TOF MS, found that, if they used genomic markers other than ITS and the focus of infection was taken into account, the species of the T. mentagrophytes complex could be distinguished.24 In this way, if the species of the T. mentagrophytes complex are added into in-house DBs, MALDI-TOF MS could be used to identify them.
There is a subgroup of genera/species (Microsporum gypseum [N. gypsea], Microsporum nanum [N. nana], Microsporum fulvum [N. fulva], Trichophyton violaceum, Trichophyton schoenleinii, Trichophyton verrucosum, Trichophyton ajelloi [A. uncinatum] and Trichophyton erinacei) in which the distance observed in the dendrogram led to inconclusive results; this could be due to the fact that they were under-represented species in the RMCABA DB. Adding more spectra for some species would enable us to improve their identification.
When the RMCABA DB was built, there were already several publications on the new taxonomic changes, but they were not conclusive. At present, it has been suggested that M. gypseum, M. nanum, M. fulvum and Trichophyton ajelloi be named N. gypsea (geophilic species), N. nana (geophilic species), N. fulva (zoophilic species) and A. uncinatum (geophilic species), in addition to other less common changes in genera and species of dermatophytes that were not included in our DB at that time. These taxonomic modifications have to do with studies based on the genome, natural reservoirs and the different proteolytic capacities for degrading the keratin of dermatophytes.6,8,9 Therefore, we suggest including explanatory notes in the report that MALDI-TOF MS generates: when species such as T. mentagrophytes and T. interdigitale are identified, they would be reported as T. mentagrophytes complex, and when M. gypseum, M. fulvum, M. nanum and Trichophyton ajelloi are identified, they would be reported as N. gypsea, N. fulva, N. nana and A. uncinatum, respectively.
The accurate identification of dermatophytes is important to confirm the aetiology of the infection and define epidemiological behaviours, and will depend not only on the experience of the technician but also on the methods used in each laboratory.
In conclusion, the success of MALDI-TOF MS depends on, among other factors, the amount of spectra included in the DB for a given species.3,13,16,18,19,23 Extending the DB with the spectra of 33 isolates, 32 of which were regional, resulted in an improved identification; previously unidentified strains were identified and scores improved, approaching concordance values consistent with a perfect identification. In agreement with other published studies, we found that designing a DB with spectra of regional isolates is a prerequisite for improving the identification of dermatophytes with MALDI-TOF MS technology, as is adding species not present in the DB provided by the manufacturer.
Conflicts of interestThe authors declare that they have no conflicts of interest.
Fundación Dr. Roberto B. Domecq [Dr Roberto B. Domecq Foundation], Centro de Educación Médica e Investigaciones Clínicas Norberto Quirno [Norberto Quirno Centre for Medical Education and Clinical Research] and BD Diagnostics System Argentina contributed with the reagents and equipment needed to conduct this study. We would also like to thank Dr. Alicia Arechavala for proofreading the manuscript and Dr. Adolfo Fox for helping with and participating in the sequencing studies of the dermatophytes entered in the database.
Red de Micología de la Ciudad Autónoma de Buenos Aires [Mycology Network of the Autonomous City of Buenos Aires]:
Susana Carnovale (Hospital Garrahan), Silvana Cataldi (Hospital Durand), Alicia Arechavala (Hospital Muñiz), Laura Dufranc (Hospital Zubizarreta), Analía Fernández (Fundación Favaloro), Norma Fernández (Hospital Clínicas) Agustina Forastiero (Hospital Británico), Claudia Garbasz (Hospital Pirovano), Liliana Guelfand (Hospital Fernández), Ricardo Iachini (Instituto Pasteur), Mónica López (Hospital Ramos Mejía), Laura López Moral (Hospital Argerich), Ivana Maldonado (Hospital Alemán), Andrea Marucco (Hospital Santojanni), Patricia Minervini (Hospital Santa Lucía), Josefina Nucci (Hospital Sardá), Paula Peña Amaya (Hospital Gutiérrez), Carolina Peloc (Hospital Álvarez), Silvia Relloso (CEMIC), Romeo Ana María (Hospital Penna) Roxana Pereda (Hospital Elizalde), Graciela Ponce (IREP), Gabriela Santiso (Hospital Muñiz), Gabriela Snitman (Hospital Quemados), Adriana Sorge (Instituto Roffo), Natalia Zalazar (Hospital Tornu), Nora Franco (Hospital Piñero).