Starting out with its discovery as small notches on fly wings, Neurogenic Locus Notch Homolog 4 (Notch4) signaling has been sparked as unique pathway implicated in cellular multiplication, differentiation, and regulation of stem cells. Its aberrant activation causes arrays of cancers including breast cancer.
ObjectivesThe aim of the present study was to evaluate the immunohistochemical expression level of Notch4 and its subcellular localization in invasive breast carcinoma. The correlation between Notch4 expression and both of clinicopathological parameters and immunohistochemical-based subtypes of studied cases was also assessed.
Methods and materialsImmunohistochemical expression of Notch4 receptor was examined in 60 specimens of paraffin-embedded sections of invasive breast cancer. Normal and hyperplastic breast tissue adjacent to carcinoma cells was also included in the study.
ResultsThere was a significant increase in the expression level of Notch4 protein in breast cancer, compared to that of normal breast tissue and hyperplastic breast lesions. Also, there was a statistical significant correlation between Notch4 expression level and tumor size, tumor grade, nodal metastasis, lymphovascular invasion, human epidermal growth factor receptor 2 (her2) status, Her2-enriched and triple negative subtypes, and Ki67. Furthermore, an inverse significant correlation was found between Notch4 expression and both of age and estrogen receptor (ER). No statistical significant correlation was found between Notch4 expression and tumor histological subtypes, Tumor infiltrating lymphocytes (TILs), and fibrosis.
ConclusionNotch4 overexpression has been implicated in breast cancer development, progression and emergence of aggressive biological phenotypes.
Empezando por su descubrimiento como pequeños surcos en las alas de la mosca, la vía de señalización del homólogo 4 del Notch del locus neurogénico (Notch4) se ha revelado como la única vía implicada en la multiplicación y diferenciación celular, y regulación de las células madre. Su activación aberrante causa una serie de cánceres, incluyendo el cáncer de mama.
ObjetivosEl objetivo del presente estudio fue evaluar el nivel de expresión inmunohistoquímica de Notch4, así como su localización subcelular en el cáncer de mama invasivo. También se evaluaron la correlación entre la expresión de Notch4 y los parámetros clínico-patológicos y subtipos con base inmunohistoquímica de los casos estudiados.
Métodos y materialesSe examinó la expresión inmunohistoquímica del receptor de Notch4 en 60 muestras de secciones parafinadas de cáncer de mama. También se incluyeron en el estudio células hiperplásicas de tejido de mama, adyacentes al carcinoma.
ResultadosSe produjo un incremento significativo del nivel de expresión de la proteína Notch4 en el cáncer de mama, en comparación con el tejido normal de la mama y las lesiones de mama hiperplásicas. De igual modo, se produjo una correlación estadísticamente significativa entre el nivel de expresión de Notch4 y el tamaño y el grado tumoral, el desarrollo de nódulos metastásicos, la invasión linfovascular, el estatus de Her2 (receptor 2 del factor de crecimiento epidérmico humano), los subtipos Her2 enriquecido y triple negativo, y Ki-67. Además, se encontró una correlación significativa inversa entre la expresión de Notch4 y la edad, y el receptor de estrógenos (ER). No se encontró correlación estadísticamente significativa entre la expresión de Notch4 y los subtipos histológicos del tumor, linfocitos infiltrantes tumorales (TIL) y fibrosis.
ConclusiónSe ha observado la implicación de la sobreexpresión de Notch4 en el desarrollo del cáncer de mama, y la progresión y aparición de fenotipos biológicos agresivos.
Breast cancer (BC) is considered the leading female cancer and the most common cause of cancer death in females worldwide.1 In Egypt, Breast is the site of one of the most frequent cancer in female, representing about 35.1% of all cancer cases among women, and contributing about 23.8% of cancer death in females.1,2
Breast cancer is classified, based on genes expression, into; Luminal (type A and B), Normal like, Her2-enriched and Triple negative breast cancer (TNBC). The two latter subtypes are associated with adverse prognosis.3
In BC, It has been proposed that a small subpopulation of undifferentiated cells exists, identified as tumor-initiating cells or cancer stem cells (CSCs). These cells are defined by their ability for self-renewal, treatment resistance and cancer relapse by depending on a variety of key signaling pathways, including the Notch pathway.4
For initiating Notch pathway, each Notch receptor of Notch family (Notch 1–4) of one cell, has to connect with one of the five transmembrane ligands (Delta-like 1,3,4 and Jagged 1, 2) of adjacent cell. Notch4, is a receptor protein encoded by Notch4 gene, located on chromosome 6. Notch4 controls several differentiation processes in cells and shares the same structure with other Notch family members. It is formed of an extracellular domain (29 EGF-like repeats), trans-membrane domain and an intracellular domain (NICD) (RAM, ANK, PEST).5
Once Notch4 receptor is activated, NICD is translocated into the nucleus, where it interacts with transcription factors to activate the targets genes.5 Activating Notch4 target genes and cross-talking with other pathways, resulting in deregulation of CSCs and tumorigenesis, through activation of c-Myc, cyclin D, mTOR, ER and Her2, decreasing apoptosis via survivin, GAS1 and AP1, increasing angiogenesis via Notch4-DLL4-VEGFR signaling and inducing epithelial mesenchymal transition (EMT) by Slug, Snail, Twist and Zeb1.4,6
In BC, sparse studies reported conflicting results regarding to Notch4 expression and its subcellular localization.7,8 Few studies attempted to prove that overexpression of Notch4 is responsible for tumorigenesis and may be a predictor for aggressive clinicopathological features, Her2-enriched and TN subtypes.8,9 Other investigators reported the association of Notch4 with Luminal subtype.10
In the present study, we tried to evaluate the potential role of Notch4 protein in initiation and progression of breast cancer. So, the expression levels of Notch4 were investigated in breast cancer cases and its subtypes and the correlation between Notch4 expression and clinicopathological parameters was analyzed.
Materials and methodsPatients and tissue specimensThis study is a cross sectional study. A total of 60 consecutive cases of selected female breast tissue, with primary invasive breast carcinomas, were collected from the received specimens at the Pathology department, Faculty of medicine, Zagazig University, Egypt, in the period from October 2018 to January 2020. The specimens were obtained either by Tru-cut biopsy (n=6), lumpectomy (n=11), or modified radical mastectomy (n=43). Normal (n=25), and hyperplastic breast tissue (n=26) adjacent to carcinoma cells were also included in the study. This study included cases of primary invasive breast carcinoma with complete clinicopathological data. Cases with incomplete data, insufficient tissue for staining or cases with a history of preoperative chemotherapy or radiotherapy were excluded from the study.
All cases of invasive breast carcinoma were histologically classified according to the WHO classification of breast tumors.11 Carcinoma in situ, lympho-vascular invasion and fibrosis, were also evaluated. Assessment of TILs was performed according to the International TILs Working Group.12 Tumors were graded according to Elstron/Nottinghfication of the Bloom-Richardson system.13 Both tumor size and the dissected lymph nodes were evaluated grossly and microscopically according to AJCC TNM Staging of breast Cancer.14 Categorization of results of ER, PR and Her2 was done according to the approved CAP recommendations.15 Ki67 index above 14% was considered high.16
The center’s protocols have been followed with respect to the treatment of patients’ data.
Immunohistochemistry protocol- 1)
ER and PR immunostaining and evaluation:
Immunohistochemical staining of ER/PR was performed by ER/PR pharmDx Kit by using EnVision system technique, Dako Autostainer (ER: Mouse, monoclonal, clone 1D5, Code.IR657, Dako, Glostrup, Denmark, Ready-to-use) (PR: Mouse, monoclonal, clone PgR 636, Code. IR068., Dako, Glostrup, Denmark, Ready-to-use). Moderate to strong nuclear staining in ≥1% of tumor cells was considered positive according to the approved CAP recommendations.15
- 2)
HER2/neu immunostaining and evaluation
Hercep Test™ kit K5207 (DAKO Cytomation, Glostrup, Denmark) was performed on all slides. Her2/neu was considered positive if circumferential membranous staining within 10% of tumor cells that is complete and intense according to the approved CAP recommendations.15
- 3)
Ki67 immunostaining and evaluation
Immunohistochemical staining of Ki67 was done (Mouse, monoclonal, clone. MIB-1, Code .IS626, Dako, Glostrup, Denmark, Ready-to-use). Ki67 index above 14% was considered high.16
- The molecular subtypes, based on the immunohistochemical evaluation of the hormone receptor expression and HER2/neu, were determined. There are 4 IHC− subtypes; Luminal A [ER+, PR+, Her2/Neu−, low Ki67], Luminal B (ER+, PR+, Her2/Neu− or+, high Ki67), Her2-enriched (ER−, PR−, Her2/Neu+, high Ki67), and Triple negative (ER−, PR−, Her2/Neu−, high Ki67).3
- 4)
Notch4 immunostaining
The immunohistochemical staining was carried out using EnVision system technique (DAKO, North America Inc, CA, USA), a polymer-enhanced two-step IHC detection system by using antibody to Notch4 (primary antibody, Notch4 C-3, clone: A-12, Code No. sc-377399, mouse monoclonal antibody from Santa Cruz Biotechnology, Inc. Europe, 0.09% sodium azide. Dilution 1:100) Kidney tissue were used as a positive control.
Interpretation of immunohistochemistryNotch4 immunnoreactivity was evaluated and scored independently in a blind manner by two investigators as that described by Wu et al, A semi-quantitative scoring system was used to evaluate both of staining intensity (0, no staining; 1+, weak staining; 2+, moderate staining; 3+, strong staining), and the percentage of stained cells (0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, >75%). The immunoreactivity score (IRS) for each case depends on multiplying scores of staining intensity and percentage of positive cells. Cases with IRS≥4 were considered to have Notch4 overexpression.17
Statistical analysisAll data were collected, tabulated and statistically analyzed using SPSS (statistical package for the social science, Chicago, Illinois, USA) version 23. Normality of data was examined by Shapiro and Kolmogrov test. Student's t-test was used to compare between two independent samples with normal distribution. Chi-square test, Fisher's exact test, Spearman's correlation coefficient and rank biserial were applied to determine the correlation between the variables. P values less than 0.05 were considered statistically significant.
Ethical approvalThis study was carried out following the Code of Ethics of the World Medical Association of studies involving humans, which was worked in 1975 by Helsinki Declaration and revised in 2000. Institutional Review Board (IRB), of the faculty of Medicine, Zagazig University, confirmed this study protocol (No. 5102). Written informed consent of participants was obtained.
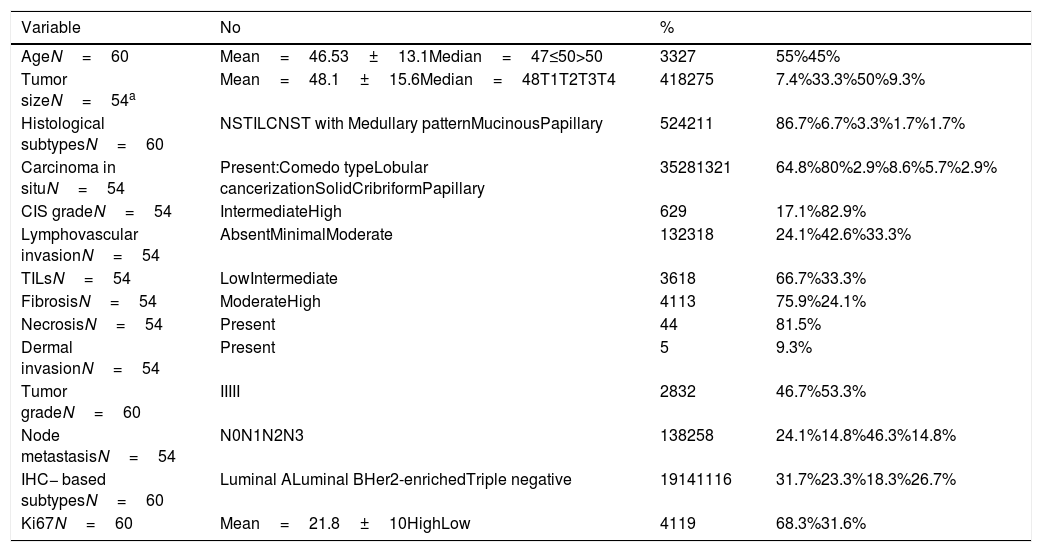
ResultsPatients’ characteristicsThe patient age ranged from 25 to72 years, with a mean and median age 46.53±13.1 and 47 years respectively. Tumor size ranged from 1 to 8cm in its greatest diameter axis, with a mean and median size of 48.1±15.6 and 48mm respectively. Most cases presented with T3 27/54 (50%), grade III, 32/60 (53.3%), N2 25/54 (46.3%), high mean ki67 proliferative index 41/60 (68.3%) and IBC-NST 52/60 (86.7%). DCIS was detected in 35 of 54 BCs (64.8%), The majority of cases 26/35 (74.3%), were associated with minor DCIS component, whereas a minority 9/35 (25.7%), had extensive DCIS (≥ 25% of the tumor area). Luminal A and Luminal B subtypes, comprised 31.7 (19/60) and 23.3% (14/60) of the studied cases respectively, while, Her2-enriched and Triple negative subtypes represented 18.3% (11/60), and 26.7% (16/60) of cases respectively. Clinicopathological characteristics are presented in Table 1.
Clinicopathological features of the studied breast cancer cases:.
| Variable | No | % | |
|---|---|---|---|
| AgeN=60 | Mean=46.53±13.1Median=47≤50>50 | 3327 | 55%45% |
| Tumor sizeN=54a | Mean=48.1±15.6Median=48T1T2T3T4 | 418275 | 7.4%33.3%50%9.3% |
| Histological subtypesN=60 | NSTILCNST with Medullary patternMucinousPapillary | 524211 | 86.7%6.7%3.3%1.7%1.7% |
| Carcinoma in situN=54 | Present:Comedo typeLobular cancerizationSolidCribriformPapillary | 35281321 | 64.8%80%2.9%8.6%5.7%2.9% |
| CIS gradeN=54 | IntermediateHigh | 629 | 17.1%82.9% |
| Lymphovascular invasionN=54 | AbsentMinimalModerate | 132318 | 24.1%42.6%33.3% |
| TILsN=54 | LowIntermediate | 3618 | 66.7%33.3% |
| FibrosisN=54 | ModerateHigh | 4113 | 75.9%24.1% |
| NecrosisN=54 | Present | 44 | 81.5% |
| Dermal invasionN=54 | Present | 5 | 9.3% |
| Tumor gradeN=60 | IIIII | 2832 | 46.7%53.3% |
| Node metastasisN=54 | N0N1N2N3 | 138258 | 24.1%14.8%46.3%14.8% |
| IHC− based subtypesN=60 | Luminal ALuminal BHer2-enrichedTriple negative | 19141116 | 31.7%23.3%18.3%26.7% |
| Ki67N=60 | Mean=21.8±10HighLow | 4119 | 68.3%31.6% |
Notch4 immunoreactivity was detected in neoplastic cells and endothelial cells lining tumor vasculature of all studied cases (100%) with the same intensity. Cytoplasmic Notch4 expression was observed in 58 of 60 cases (96.7%), while simultaneous nuclear and cytoplasmic Notch4 expression was detected in only 2 cases representing 3.3% of studied cases. According to the Notch4-IRS, Notch4 overexpression was detected in 33 of 60 BC cases (55%), with IRS≥4 (range 4–12). Among them, 72.7% (24/33) of cases had diffuse and moderate Notch4 staining intensity, while 9/33 (27.3%) were diffuse and strong. Notch4 overexpression was higher in both TN (75%) and Her2-enriched (72.7%) compared to Luminal (39.4%). The remaining, 27 cases (45%), showed low Notch4 expression (range 2–3), with either focal, 8/27 (29.7%) or diffuse (but less than 75%), 19/27 (70.3%), weak Notch4 immunoreactivity (Fig. 1). Notch4 overexpressing cases showed heterogenous immunoreactivity with increased Notch4 staining intensity at the invasive edges, where malignant cells interface with stroma (Fig. 2).
The majority, 20/25 (80%), of normal breast tissue and most, 15/26 (57.7%), of hyperplastic lesions, showed a predominantly low level of Notch4 expression. There was a significant increase in Notch4 expression in BC compared to that of normal (P=0.005) and hyperplastic breast lesions (P=0.021) Table 2. Regarding to DCIS, Notch4 overexpression was detected in 20/35 (57.1%), including comedo 16/29 (55.2%), lobular cancerization 1/1 (100%), solid 1/3 (33.3%), cribriform 1/2 (50%), and papillary 1/1 (100%) subtypes. Each case of DCIS showed the same immunostaining intensity as that of its invasive component within the same tumor.
Notch4 immunreactivity evaluation in breast cancer and adjacent tissues (normal and hyperplastic leisons), test association by Fisher's exact test.
Correlation between Notch4 and clinicopathological parameters were assessed and presented in Table 3: There was a statistical significant inverse correlation between Notch4 expression and both patient age (r=−.263, P=.043) and ER (r=−.622, P<.001) Fig. 3A. Statistically significant correlation was found between Notch4 expression and histological grade (r=.387, P=.002), DCIS grade (r=.348, P=.040), pT (r=.342, P=.018), pN (r=.366, P=.006), lymphovascular invasion (r=.303, P=.026), Her2 status (r=.369, P=.004) and Ki67 index (r=.719, P<.001) (Fig. 3B). Regarding to subtypes, Notch4 expression was significantly correlated with triple negative (r=.271, P=.036), and Her2-enriched (r=.261, P=.044) subtypes, without significant difference in the correlation between them (P=.791). There was no statistically significant correlation between Notch4 expression and histological subtypes, TILs and fibrosis.
Correlation between Notch4 overexpression and clinicopathological parameters:.
| No (%) | Notch4Low expression | Notch4Overexpression | r | P | |||
|---|---|---|---|---|---|---|---|
| No | % | No | % | ||||
| Age | t .039* | ||||||
| Mean | 49.7±12 | 42.4±14 | |||||
| <50 | 33 (55%) | 11 | 33.3% | 22 | 66.7% | −.263 | .043* |
| ≥50 | 27 (45%) | 16 | 59.3% | 11 | 40.7% | ||
| pT | |||||||
| T1 | 4 (7.4%) | 3 | 75% | 1 | 25% | ||
| T2 | 18 (33.3%) | 13 | 72.2% | 5 | 27.8% | .342 | .018* |
| T3 | 27(50%) | 9 | 33.3% | 18 | 66.7% | ||
| T4 | 5 (9.3%) | 2 | 40% | 3 | 60% | ||
| Histological subtypes | |||||||
| NST | 52 (86.7%) | 23 | 44.2% | 29 | 55.8% | .080 | |
| Other | 4 (6.7%) | 2 | 50% | 2 | 50% | .545 | |
| ILC | 2 (3.3%) | 1 | 50% | 1 | 50% | NS | |
| Medullary like papillary | 1 (1.7%) | 0 | 0% | 1 | 100% | ||
| Mucinous | 1 (1.7%) | 1 | 100% | 00 | 00 | ||
| LIN | |||||||
| Ly 0 | 13 (24.1%) | 9 | 69.2% | 4 | 30.8% | .303 | .026* |
| Ly 1 | 23 (42.6%) | 12 | 52.2% | 11 | 47.8% | ||
| Ly 2 | 18 (33.3%) | 6 | 33.3% | 12 | 66.7% | ||
| Ly 3 | 00 | 00 | 00 | 00 | 00 | ||
| TILs | |||||||
| Low | 36 (66.7%) | 21 | 58.3% | 15 | 41.7% | .236 | .086 |
| Intermediate | 18 (33.3%) | 6 | 33.3% | 12 | 66.7% | NS | |
| High | 00 | 00 | 00 | 00 | 00 | ||
| Fibrosis | |||||||
| Moderate | 41 (75.9%) | 20 | 48.8% | 21 | 51.2% | .043 | .756 |
| High | 13 (24.1%) | 6 | 46.2% | 7 | 53.8% | NS | |
| Tumor grade | |||||||
| II | 28 (46.7%) | 17 | 60.7% | 11 | 39.3% | .387 | .002* |
| III | 32 (53.3%) | 10 | 31.3% | 22 | 68.8% | ||
| Grade of CIS | |||||||
| Grade II | 6 (17.1%) | 3 | 50% | 3 | 50% | .348 | .040* |
| Grade III | 29 (82.9%) | 12 | 41.4% | 17 | 58.6% | ||
| pN | |||||||
| N0 | 13 (24.1%) | 9 | 69.2% | 4 | 30.8% | .366 | .006* |
| N1 | 8 (14.8%) | 5 | 62.5% | 3 | 37.5% | ||
| N2 | 25 (46.3%) | 7 | 28% | 18 | 72% | ||
| N3 | 8 (14.8%) | 2 | 25% | 6 | 75% | ||
| IHC− subtypes | |||||||
| Hormonal: Luminal A | 19 (31.7%) | 16 | 48.5% | 3 | 9.1% | .352 | .006* |
| Luminal B | 14 (23.3%) | 4 | 12.1% | 10 | 30.3% | −.369 | .004* |
| Her2− enriched | 11 (18.3%) | 3 | 27.3% | 8 | 72.7% | .261 | .044* |
| TN | 16 (26.7%) | 4 | 25% | 12 | 75% | .271 | .036* |
| ER | |||||||
| Negative | 27 (45%) | 7 | 25.9% | 20 | 74.1% | −.622 | <.001** |
| Positive | 33 (55%) | 20 | 60.6% | 13 | 39.4% | ||
| Her2 status | |||||||
| Negative | 35 (58.3%) | 17 | 48.6% | 18 | 51.4% | .369 | .004* |
| Positive | 25 (41.7%) | 6 | 24% | 19 | 76% | ||
| KI67 | t <.001** | ||||||
| Mean | 8.4±7 | 24.6±8 | .719 | ||||
| High | 41 (68.3%) | 11 | 26.8% | 30 | 73.2% | <.001** | |
| Low | 19 (31.6%) | 16 | 84.2% | 3 | 15.8% | ||
ER+=PR+ in all 60 cases.
t: Student's t-test.
In this study, Notch4 immunoreactivity was detected in all studied BC cases, both in neoplastic cells and vascular endothelial cells. Notch4 immunoreactivity was mainly cytoplasmic and simultaneously cytoplasmic and nuclear in only two studied cases. These results confirm the observation of Wang and colleagues, who found cytoplasmic Notch4 expression in all their studied cases.8 However, Yao et al. and Speiser et al. demonstrated more cytoplasmic Notch4 expression than membranous,7 and simultaneous cytoplasmic and nuclear expression of their studied cases9.
Cytoplasmic localization of Notch4 may represent functional recently synthesized protein, while membranous localization may represent inactive receptor. However, nuclear Notch may represent an activated receptor, which is hardly detected by simple immunohistochemical technique, due to its rapid degradation.7,18 The mechanisms that control nucleo-cytoplasmic shuttling of NICD are not clear. It is documented that Mdm2, the PI3K-AKT pathway and 14-3-3 regulatory proteins are involved in NICD degradation and phosphorylation.19,20
In the present study, Notch4 overexpression was observed in 55% of BC cases. These results are inconsistent with the previous studies that revealed either higher levels (81.5%),21 or lower levels (39%),8 of Notch4 overexpressing cases. The differences between these studies could be attributed to different criteria of Notch4 evaluation or experimental methods sensitivity, or the difference of clinicopathological features and genetic bases of the studied cases.
The finding of heterogenous Notch4 immunoreactivity of malignant cells, was previously reported by Stingl et al. who attributed it to the possible diversity of neoplastic cell origin, from breast epithelial stem cells or their progeny.22 Also, increased staining intensity at the invasive edges of tumors, observed in our study, was previously reported by Zhou et al., who reported deviation of epithelial cells toward a mesenchymal trans-differentiation in the EMT dependent Notch4.6
In the present study, endothelial cells of vessels tumor showed Notch4 overexpression compared to normal vessels in adjacent normal tissue and followed the same expression of neoplastic cells. In contrast to this result, Speiser and colleagues, found strongly immunoreactive for Notch4 in endothelial cells of all 29 studied. In breast cancer, Notch4-DLL4-VEGFR signaling system is a major activator of angiogenesis.4,9
In this study, There was a statistical significant increase in Notch4 expression in BC, compared to normal breast tissue, (P=0.005). This result is consistent with other studies.6,21 On the other hand, Ma et al., could not detect Notch4 in normal breast tissue,23 while Mittal et al., could not find significant change in the level of Notch4 expression between normal and cancer tissues.24
In normal breast tissue, Notch pathway plays an essential role in enhancing mammary gland homeostasis by cell-fate decisions regulation such as self-renewal of adult stem cells and differentiation of progenitor cells, resulting in cell survival and cell proliferation by activation of target genes and cross talk with other pathways such as, Hes, ER, Wnt and Hg.4 However, in aberrant Notch receptors activation, the mammary stem cells could be disrupted, with the release of breast cancer stem cells.4 The molecular mechanisms of this aberrant activation of Notch signaling may be associated with mutations of Notch receptors genes,8 or lower expression of Notch negative regulators in cancer cells such as FBXW, and Numb, or loss of differentiation inducing factors, including E74 like factor 5 and ring finger protein.4
Regarding to Notch4 expression in hyperplastic lesions and DCIS, our results suggest that activation of the Notch4 pathway in BC may occur as early as hyperplasia and DCIS. This confirms the observation of previous studies who suggested that activation of the Notch4 pathway may be a primary trigger in the BC onset.7,24
In the present study, there was a statistical significant correlation between Notch4 expression and young age (r=−.263, P=0.043). In contrast to this result, Wang and colleagues, found no correlation among Notch4 expression and patients’ age.8 This difference may be attributed to participation of other factors such as sample size, tumor grade or its subtype.
Regarding to tumor size, there was a statistical significant correlation between Notch4 expression and tumor size (r=0.342, P=0.018). This result is consistent with one study of Wang et al.,8 and inconsistent with the other Touplikiot et al.10
With regard to Notch4 expression in histological subtypes, there was no statistical significant correlation between Notch4 expression and histological subtypes (r=0.080, P=0.545). These results are consistent with a previous study done by Ma et al.23 On the other hand, Rizzo et al. and Touplikiot et al., found higher level of Notch4 in ILCs, which couldn’t be demonstrated in this study due to the small number of the studied ILCs cases.10,21
In the present study, there was a statistical significant correlation between Notch4 expression and lymphovascular invasion (r=0.303, P=0.026). This comes in agreement with a previous study done by Wu and colleagues, who reported that Notch4 overexpression denoted the presence of lymphovascular invasion in cancer colon.17
In this study, no statistical significant correlation between Notch4 expression and both TILs (r=0.236, P=0.086), and fibrosis (r=0.043, P=0.756) could be detected. Notch may play a role in tumor induced immunosuppression. Myeloid-derived suppressor cells in BC can activate Notch signaling in cancer cells and promote CSC capacity through IL6/STAT3 and Nitric Oxide/Notch cross talk signaling.25 Also, several studies confirmed the role of Notch signaling in the development of fibrosis in different organs.26
With regard to tumor grade, there was a statistical significant correlation between Notch4 expression and tumor and DCIS grade (r=0.387, P=0.002), (r=.348, P=0.040) respectively. In contrast to this results, Touplikiot et al. found increased Notch4 level in well-differentiated tumors without statistical significance.10 In our study, increased Notch4 level in high grade tumors may be a reflection of a genetic background of studied cases or other participating parameters, or may denote the role of Notch4 in tumor progression via dedifferentiation.
According to lymph node metastasis, there was a statistical significant correlation between Notch4 expression and nodal stage (r=0.366, P=0.006). This finding was consistent with one study,8 and inconsistent with others.7,10 Increased level of Notch4 in tumors with greater lymph node metastasis in this study, may confirm its role in the process of tumor invasion and metastasis. Tumor metastasis is promoted by Notch4 through acquiring mesenchymal features achieved by Snail, Zeb1 and Notch4-Slug-Gas1 circuit that has been documented in promoting metastasis, chemoresistance and decreasing apoptosis.6
Regarding to IHC-subtypes, Notch4 expression was correlated with IHC-subtypes (r=.352, P=.006) more toward negative hormonal expression. Notch4 expression was inversely correlated with ER (r=−0.622, P<.001). Similar results were obtained by previous studies.7,8,21 Rizzo et al. and Yao et al. revealed that estradiol decreases Notch transcription activity in BC cells.7,21 On the other hand, Calaf and Roy, reported that E2 and parathion, either combined or alone, led to Notch pathway activation in MCF-10F cell lines.27 Therefore, combinations of antiestrogen therapy and Notch inhibitors may be more efficient in ERα+BCs.21,27
Regarding to Her2-enriched subtype of BC, the present study found a statistical significant correlation between Notch4 expression and Her2-enriched subtype of BC (r=0.261, P=0.044). Significant correlation was found also between Her2-status and Notch4 expression (r=.369, P=0.004). Our result confirms the finding of Magnifico et al.28 On the other hand, Wang et al. did not find such significant association between Notch4 and Her2 status.8 Both Notch4 and Her2 pathways are implicated in the BC progression and CSCs regulation. Her2 activity is regulated by Notch4. Activation of Notch induces the transcription of Her2 that in turn, drives stem cell self-renewal.28
Regarding to TN subtype, this study found a statistical significant correlation between Notch4 expression and TNBC (r=0.271, P=0.036) with the highest Notch4 expression. Similar observation was reported in many studies.6,8,9,29 Metastatic TN are quite associated with Notch4 signaling.30
In the present study, There was a statistical significant correlation between Notch4 expression and Ki67 (r=0.719, P<.001). A similar result was obtained by Yao et al, who detected an association between cytoplasmic Notch4 and Ki67 expression, suggesting that tumors with high Notch4 expression have higher proliferation rates.7
ConclusionOverexpression of Notch4 may play an important role in the development of breast cancer and its progression by promoting cell proliferation, vascular invasion and metastasis. Also, overexpression of Notch4 has been implicated in emergence of TN and Her2-enriched subtypes, compared to Luminal subtype. So, inhibition of Notch4 signaling in breast cancer using Notch4 antagonists, may be an effective strategy to develop a targeted therapy.
FundingThe authors declare that they have received no funding for this project.
Conflict of interestThe authors declare that they have no conflict of interest.