We review the microbiological aspects of COVID-19 infection and present the microbiological studies that should be performed in forensic cases. We describe the taxonomic characteristics of the virus, its relationship with the coronaviridae family and its genetic structure. We briefly present the clinical and pathological characteristics of COVID-19 infection, as well as the co-infections that could be associated with this virus. In the laboratory, PCR is a first-choice technique in the acute phase of the infection, together with antigen and serological studies. Finally, we describe the main objectives of microbiological studies in the deceased in relation to the COVID-19 pandemic, as well as the main post-mortem microbiological analysis to be carried out in the medico-legal context. The microbiological analysis should aim to detect both SARS-CoV-2 and coinfections, which may also contribute to the cause of death.
En este artículo se revisan los aspectos microbiológicos de la infección COVID-19 y se presentan las recomendaciones sobre los análisis que deben realizarse en casos forenses. En primer lugar, se analizan las características taxonómicas del virus, su relación con la familia coronaviridae y su estructura genética. Se presentan brevemente las características clínicas y patológicas de la infección COVID-19 así como las coinfecciones que pueden asociarse a este virus. En el diagnóstico de laboratorio se describe la PCR, técnica de elección en la fase aguda de la infección; los estudios antigénicos y los serológicos. Finalmente se detallan los principales objetivos para los estudios microbiológicos en fallecidos en relación a la pandemia COVID-19 y se describen los principales análisis microbiológicos post- mórtem a realizar en fallecidos en el ámbito forense. Los estudios microbiológicos deben estar dirigidos tanto a la detección del SARS-CoV-2 como de las coinfecciones, que también podrían contribuir a la causa de muerte.
Coronavirus (CoV) is the common name for members of the Orthocoranavirinae sub-family. Taxonomically they belong to the Coronaviridae family, Nidovirales order. These viruses are widely distributed and were found to be human pathogens in the 1960s. They are zoonotic viruses that can be transmitted between animals and humans, so they have a major economic and social impact. The name coronavirus results from the structure of the virus when seen under an electron microscope, as it seems to have a sort of crown on its surface, similar to the appearance of the sun.1
There are four types of coronavirus, differentiated by the sequence of their proteins: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. The first two infect mammals and may have their reservoir in bats, while the Gammacoronavirus genus includes all of the avian coronaviruses. The Deltacoronavirus affect mammals as well as birds.2
Seven types of coronavirus infect human beings, and before the outbreak of SARS-CoV in 2003 they were considered to be the cause of mild and self-limiting respiratory infections. The human coronaviruses now known are: coronavirus 229E (HCoV-229E), coronavirus OC43 (HCoV-OC43), SARS CoV, coronavirus NL63 (HCoV-NL63), human coronavirus HKU1 (HCoV-HKU1), Middle East Respiratory Syndrome coronavirus (MERS-CoV) and Wuhan coronavirus o SARS-CoV-2. Four of these seven viruses (HCoV-229E, HCoV-OC43, HCoV-NL63 and HCoV-HKU1) specifically affect the human species and cause from 15% to 30% of respiratory tract infections every year, with very severe symptoms in new born babies, the elderly and people with underlying diseases. They mainly affect the lower respiratory tract.3,4
The other three coronaviruses (SARS-CoV, MERS-CoV and SARS-CoV-2 of 2019) are highly pathogenic and cause severe infections of the lower respiratory tract, leading to acute difficulty in breathing and manifestations outside the lungs. The SARS outbreak in 2003 led to a rethink of these viruses and their role in human infections.5,6 Ten year after this first outbreak, another one emerged in the Arabian peninsula (MERS), from where it propagated sporadically to the rest of the world.7–9 The new SARS-CoV-2 coronavirus is the cause of the current pandemic, and it has given rise to an unprecedented medical and economic crisis in the modern age.
The complete sequence of the SARS-CoV-2 genome, as determined by mass sequencing, established significant differences with previous types of coronavirus that had caused outbreaks (SARS and MERS). Detailed analysis of the sequence10 made it possible to establish 96.2% homology with a bat coronavirus (Bat-SARS RaTG13), so that it is included, together with the said virus, in a different lineage of the subgenus of Sarbecovirus.11 Although the exact sequence of transmission between bats (the main reservoir) and other mammals cannot be established, its zoonotic origin seems indisputable.
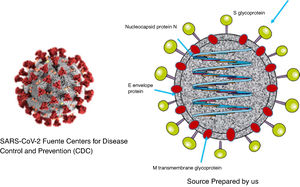
As was the case with SARS and MERS, SARS-CoV-2 is a virus with a two layer lipid envelope of approximately 100–160 nanometres in diameter. The envelope has spicules that project outwards and are composed of glycoprotein S trimer.
The viral genome is a simple sequence with positive polarity. The 5′ end of the same contains the genes that code for regulatory proteins that will give rise, among others, to the protease, the RNA-dependent polymerase RNA and the helicase. The genes which code for the structural proteins (S [spike protein], E [envelope], M [membrane] and N [nucleocapsid]) are located in the 3′ end.12. Additionally, the virus codifies non-structural proteins1,13 (Fig. 1).
Protein S forms structures that project from the viral envelope. It contains the domain for binding to the receptor of the cells which it infects (RBD), so that it is therefore the protein that determines the tropism of the virus. It is composed of two different domains: domain S1, which is responsible for binding to the receptor, and domain S2, which is responsible for the fusion with the cellular membrane.14
The sequence and analysis of SARS-CoV-2 protein S indicate that, as was the case with SARS-CoV-1, it uses the angiotensin converting enzyme 2 (known by its initials ACE2) as receptor for entry into the host cell.15 ACE2 is located on the surface of a wide range of cells in the mucous membranes, lungs, arteries and intestines, etc., where it converts angiotensin I into angiotensin II, increasing its vasoconstrictive action. The virus uses this molecule to enter into the cell, where the cellular ribsosomes use the viral RNA as messenger RNA to synthesize the virus proteins. This, together with the viral replicase, makes it possible to make multiple copies of the virus that favour its dissemination.
N protein is contained in the virion associated with viral RNA, and it plays an important role in the replication of the virus and the assembly of new viral particles. M protein is the most abundant, and it is responsible for the final shape of the virion. E protein is of small size and is found in small amounts on the surface layer. The non-structural proteins play important specific roles in the virus replication process.
The fact that SARS-CoV-2 has reached human beings from an animal origin means that there is a high probability of outbreaks by similar viruses in the future, as this type of virus is still circulating in the animal population. It is therefore a priority to know its characteristics (transmissibility, pathogenicity and rate of evolution, etc.) as these will condition its propagation and determine the extension of the pandemic. It is also of special interest to discover whether SARS-CoV-2 may be seasonal, as are the majority of the coronaviruses that infect humans.
COVID-19 diseaseClinical manifestationsThe binding of SARS-CoV-2 to the ACE2 receptor, which is located among other places in type I and II alveolar cells, may cause lesions at the level of these cells, particularly in those which have a large amount of the receptor. This damage may trigger a massive release of cytokines, which, together with the capacity of the virus to evade the immune response, plays an important role in the pathogenesis and severity of the disease.16
The clinical manifestations in cases of COVID-19 vary very widely, from asymptomatic cases to ones with septic shock, pneumonia and multiple organ failure. The disease is classified in different types according to its severity. The most common symptoms are fever, fatigue, a dry cough and diarrhoea. After an incubation period of from 5 to 14 days, 80% of cases appear with mild localised symptoms in the upper airways and accompanied by non-specific symptoms such as fever, asthenia, anosmia, ageusia and myalgia. Some patients may deteriorate quickly and evolve into a more serious clinical picture, with dyspnoea, taquipnea, a fall in saturation and bilateral alveolar infiltrates, which may require admission to hospital. A variable percentage of the total number of cases may in turn require admission to intensive care units (ICU) due to severe pneumonia, severe respiratory distress syndrome, septic shock or multiple organ failure.17 These patients have severe dyspnoea, taquipnea (> 30/min), SpO2 < 93%, PaO2/FiO2 < 300 and/or a 50% increase in pulmonary infiltrates in 24 h. or 48 h. The most common extrapulmonary complications are cardiovascular, hepatic and renal. Lymphopenia and very high levels of dimer-d, ferritin, urea and creatinine are characteristic in laboratory parameters. Multiple organ failure occurs due to an excessive response of the patient’s immune system. The Chinese CDC suggests that there may be up to 49% mortality in critical patients, chiefly because of the presence of comorbidities such as diabetes, respiratory disease, cardiovascular disease, hypertension and oncological complications.18
The general mortality in the Wuhan series amounted to 5.8%,16 although different studies question this figure. In any case, the real mortality figures are hard to calculate due to the highly heterogeneous nature of some data.
PathologyLungsMacroscopically: increased in size, with opaque pleura. The cut surface shows consolidation and/or oedema. In the case of secondary bacterial infection purulent exudate may be added.19
Microscopically: in early phases of the infection intraavleolar oedema is observed, together with hyperplasia of type II pneumocytes, giant cells, focal inflammation and generally no presence of hyaline membranes; in more advanced phases of the infection diffuse alveolar damage is observed, with hyaline membranes, fibrin exudates, interstitial lymphocytic infiltrate (pneumonitis), presence of giant cells and atypical desquamated pneumocytes in the alveolar space with prominence of nucleoli. The lymphocytes correspond to type T (principally CD4, and less CD8). Recently viral particles have been detected in the lungs of patients affected by COVID-19,20 although other authors have not detected them.21–24 Some authors describe the presence of microthrombi in the alveolar capillaries. If there is bacterial coinfection, abscesses may be added to the histological signs.20
HeartMacroscopically: it may show areas of haemorrhage and signs of infarct.
Microscopically: endothelial damage may be observed, with accumulation of inflammatory cells in relation with the endothelial cells (lymphocytic endothelitis) and apoptosis in the myocardium. Histological signs may also be found of lymphocytic myocarditis. The virus has been detected in the cytoplasm of the affected endothelial cells.
Other organsThe development of ischemia has been described (macroscopically) and/or lymphocytic endothelitis (microscopically) in the colon, small intestine, mesenterium, kidneys and liver.25
Cases of encephalitis in patients with COVID-19 have been described very recently, including necrotizing haemorrhagic encephalitis.26,27
Coinfections in COVID-19 diseasePneumonia is one of the main causes of hospital admission. This condition may be caused by bacteria, virus, fungi or parasites. A single microorganism is usually responsible for the symptoms, although this is not always the case. Current diagnostic techniques have made it possible to find that in many cases more than one microorganism is present in these pathologies, and that when several are present this aggravates the symptoms, complicating patient recovery. This is applicable not only to traditional pneumonia, as it also occurs with COVID-19.
Bacterial and fungal coinfectionsHospitalised patients, and especially those in an ICU, are at high risk of developing nosocomial infections or pneumonia associated with mechanical ventilation, as well as secondary coinfections that are not associated with a respiratory manifestation (such as a urinary infection). Bacterial and fungal infections are common complications of viral pneumonia, especially in critical patients. Nevertheless, few studies of infections of this type have been conducted in patients with COVID-19, and the majority of published works do not include a prognostic analysis, even when secondary infection has been found in 50% of non-surviving patients with COVID-19.28
A review of the existing literature indicates that 8% of all patients admitted to hospital suffered fungal or bacterial coinfections. The most frequent form of coinfection is bacteraemia, and the microorganisms found are gram-positive bacteria (Staphylococcus aureus) as well as gram-negative strains (Acinetobacter baumannii, Klebsiella pneumoniae); atypical bacteria were detected in respiratory samples (Legionella sp, Chlamydophila pneumoniae).29
Coinfection by Aspergillus was also found in several subjects with respiratory distress, and it shows the need for doctors treating patients with this pathology, even if it is caused by COVID-19, to consider the possibility of invasive pulmonary aspergillosis and, as a result of this, to process respiratory samples with specific analyses that make it possible to detect coinfection.30
Although it may be hard to distinguish between bacterial or fungal infection and the existing viral pneumonia on the basis of symptoms and radiology, microbiological examination – especially sputum culture – can offer extremely valuable information, on condition that the laboratory staff maintain suitable protective measures while handling these samples with high infectious potential.
The percentage of patients with COVID-19 in which coinfections of this type have been detected is small in comparison with other coronaviruses. However, this may be explained by the fact that high levels of antibiotics have been administered during this pandemic, and broad spectrum antibiotics have often been used; nevertheless, multi-resistant microorganisms have not been identified very often.
A high proportion of the bacterial coinfections mentioned in the literature on coronavirus are associated with medical treatment, including bacteraemia and infection associated with mechanical ventilation. Infection control measures must be rigorously applied under these conditions, and antimicrobial drugs must be prescribed, given that antibiotics play an important role in treating COVID-19. Combined therapy with hydroxychloroquine azithromycin has been used empirically to treat patients with pneumonia.
Viral coinfectionsThe aforesaid symptoms to classify a patient as possibly COVID-19 (fever, asthenia, dry cough, myalgia, headache, dizziness, abdominal pain, diarrhoea, nausea and vomiting) are shared by many acute respiratory infections, including viral ones.31,32 Although radiological images may serve as guidelines, they cannot completely rule out the presence of another virus in patients with the suspicion of infection by SARS-CoV-2.
Several studies have therefore been performed to detect viral coinfections in patients with the symptoms of COVID. Thus Lin et al.,33 in a work that included 186 individuals with symptoms compatible with COVID-19, found coinfections with other respiratory viruses in 6 patients (3.2%), and they found that 18 other patients had one or more respiratory viruses other than SARS-CoV-2 (rhinovirus, respiratory syncytial virus, adenovirus, influenza A and B, parainfluenza 2 and 3, coronavirus HKU1 and metapneumovirus) which were the causes of the observed symptoms.
Similar data have been found in other series undertaken in different countries. In the United States, a study that included more than 5000 patients in the area of New York found a rate of respiratory virus coinfection of 2.1%.34
Coinfections of SARS-CoV-2 and other coronaviruses have been described,35 with metapneumovirus36 and influenza virus.37,38 All of these data underline the importance of maintaining the suspicion of possible coinfection and not excluding any respiratory virus (including SARS-CoV-2) when the presence of another virus is detected. This fact is especially important, given that the possibly seasonal nature of the coronavirus that has caused the current pandemic is unknown.
The role that the immunological state of the host may play in the development of SARS-CoV-2 infection is unclear, and given that there are more than 37 million people infected with HIV in the world, it is important to know how this infection may affect the development of COVID-19. Additionally, it has been suggested that antiretroviral drugs be used as an alternative therapy, and remdesivir has now been presented as a promising option for treatment. Together with some data from China,39,40 a study undertaken in our country41 seems to indicate that these patients evolve in a similar way to the general population, although it is true that the number of cases is insufficient to consider these conclusions as definitive.
Comorbidities aggravate the prognosis of COVID-19. Elderly patients, those with diabetes, severe respiratory complications, hypertension, cardiovascular diseases, altered leukocyte/lymphocyte counts in the blood, etc., are at higher risk of a poor clinical outcome. Viral, bacterial or fungal coinfections are another factor that has to be considered an additional risk.
Microbiological diagnosis of SARS-CoV-2It is important to swiftly identify COVID-19 by laboratory tests so that patients infected with SARS-CoV-2 can be identified, isolated and treated as soon as possible, to restrict the transmission of the virus as well as to free up space in the emergency department or evaluate the condition of medical staff exposed to the virus.
The sample of choice for this diagnosis is a nasopharyngeal swab (NPS), given that the viral load is higher in the nasal fossa than it is in the oropharynx. The most useful technique is the detection of SARS-CoV-2 nucleic acids in the nasopharyngeal exudate sample, as the virus can be detected from the first phases of infection. Correct taking of the sample is fundamental to prevent false negatives; for this the patient’s head has to lean backwards, introducing the swab through the nasal fossa to the nasopharynx, rotate the swab energetically and then withdraw it. Pharyngeal samples may also be used, of endotracheal aspirate, bronchoaspirate and bronchoalveolar washing.42
Real-time PCR and other molecular analysis techniquesNucleic acid amplification techniques, and particularly RT-PCR (reverse transcription polymerase chain reaction, PCR from here on), is considered to be the technique of choice for acute phase COVID-19 disease. In patients with symptoms of infection these techniques have a high positive predictive value (PPV).
Although the majority of assays used are commercial, some reference laboratories use in house tests. The regions these assays detect include ORF1ab and regions which code S and N proteins. The selection of the said regions has the aim of maximising the specificity of the amplification to prevent crossed reactions with other related coronaviruses. Some assays include the detection of the gene which code for E protein as screening, as this protein is common to other SARS group coronaviruses, so that it would give positive results for other related coronaviruses such as 2003 SARS-CoV and bat SARS-CoV. Neither of these two viruses is circulating, and in the case of the second one, it does not infect human beings. Some of these kits analyse two or more of the said regions in multiplex format, i.e., in the same reaction, using a control that is added before extraction to verify the effectiveness of the whole process – extraction and amplification – and the absence of inhibitors in the reaction.
The outstanding commercial PCR tests include those by Altona, Thermofisher, Seegene, Genesig, Biomerieux, Bio-Speedy and Bioeksen Turkey, as well as the ones that are made in Spain, such as Vircell, Genomica or Genetics PCR Solution. There are also other formats, such as PCR-microarray hybridisation, although real-time PCR is the most widely used technique. Unpublished data suggest that techniques have different sensitivities, although in general the detection limit is usually 10 copies of viral genome per reaction.
PCR technique has also made it possible to know that in the first days with symptoms, and probably in the presyndrome phase too - infected individuals have a high viral load (from 104 to 108 genome copies/mL per nasopharyngeal or saliva sample).43 When the course of the infection is mild, the viral load peak occurs during the first 5–6 days after the start of the symptoms, and it disappears on around the tenth day. Although viral may exist in these samples after this day, the viral load is from 100−1,000 times less, indicating that the virus is hardly transmissible during this time. On the contrary, in those with severe infection the viral load is up to 60 times greater than it is for those with mild symptoms,44 and viral excretion may last for longer. One study showed that patients who had died due to severe infection by COVID-19 continued to give a positive PCR until death, and it did not become negative at any time.45 Respecting viral transmissibility, it is thought that in milder cases this commences one or two days before the start of symptoms, and that it lasts for 5 or 6 days. In more severe cases the transmission period would be longer and more intense, due to the greater viral load,43 and some recent studies even describe the possibility of an asymptomatic period during which the virus is transmissible of up to 6 days.46
PCR interpretation must always take place with the clinical context, especially when it is negative. False negatives may be due mainly to inappropriate sample taking or transport, or a low viral load at the time the sample was taken, so that it was below the sensitivity limit of the technique.
Antigen detection techniquesThe use of immunochromatography techniques to detect the SARS-CoV-2 antigen in respiratory samples (mainly in nasopharyngeal exudate) could be the first step in detecting the pathogen in symptomatic and even asymptomatic patients during the first moments of infection, given that the viral load is high at these times. Moreover, the simplicity of the technique as well as the fact that no specific equipment is used makes it an ideal candidate.43
However, almost no kit with these characteristics exists in the world market. There is only the BIOEASY™ Diagnostic Kit for Novel Coronavirus (COVID-19) Ag Test Kit, which was purchased by the Ministry of Health and Consumption and evaluated prior to use by the medical authorities for validation. The first kit studied – a conventional immunochromatograph with simple visual reading using colloidal gold – as well as the second version that was sent, which used fast immunofluorescence, showed very low levels of sensitivity (<30%), so that in no case could they be employed as useful diagnostic tools.42
Antibody detection techniquesDifferent tests with variable characteristics of sensitivity and specificity exist for the study of antibodies, and in some cases their validations have been limited by the urgent nature of the pandemic. Some of them have now been independently evaluated to unify criteria, as well as to validate these tests clinically.42 In Spain, specifically the SEIMC and the CNM are gathering information to make reliable data available at a national level on the sensitivity and specificity of these kits, and these data will be available in the near future.47
For serological study methods to detect humoral response to S protein and N protein are used, as both have been shown to be strongly immunogenic. S protein binds to a cellular receptor, the angiotensin converting enzyme ACE2, in the case of SARS-CoV-1 as well as in the case of SARS-CoV-2. Some anti-S antibodies, including those against RBD, have a neutralising activity, also it has yet to be determined its real relative proportion in comparison with the total of anti-SARS-CoV-2 antibodies during infection by the virus.48
Once infection has occurred and after the appearance of the symptoms, after approximately 5 days IgM and IgA appear; subsequently, at 14 days IgG appears (10−18 days).45,50 Seroconversion for total antibodies takes place at day 11.47,49
Some tests that combine IgM and IgG may offer results that are hardly definitive, as there may be potential crossed reactivity with other antibodies against other coronaviruses, as well the frequent low level of specificity of IgM.50
Given that a variable response has been detected to both immunoglobulins during the course of the disease, a negative result for IgM and for IgG would not rule out the patient being infected by SARS-CoV-2, especially if they are immunodepressed. Thus although antibody study is a useful tool for estimating the proportion of individuals who have been exposed to SARS-CoV-2, it is less useful in the diagnosis and clinical evaluation of a patient except under highly specific circumstances. One example of these would be the detection of patients infected with PCR-negative, particularly those that present results with a viral load below the detection limit of the PCR technique, or when it is not possible to take samples from the lower respiratory tract.51
Immunochromatography. Point of care (POC)These are known in many cases as fast tests, given that the results can be read in 10−15 min. They are actually useful to detect the response of IgM and IgG antibodies, so that sufficient time must have passed after infection for the said antibodies to have been produced.47 At the present time in Spain there are more than 60 kits with the CE mark for antibody detection. One of their advantages is that they can be used with peripheral blood (even by capillary puncture), and not only in serum or plasma. However, other ELISA or chemoluminescence techniques are more sensitive and specific.
Some of the kits in the market detect the present of total IgG and IgM antibodies against different antigens of the virus, while others detect the presence of IgM and IgG antibodies separately.
The kit purchased at first by the Ministry of Health and Consumption for screening detected IgM and IgG together against the S protein (COVID-19 IgG/IgM Rapid Test Cassette, Zhejiang Orient Gene Biotech Co Ltd, Huzhou, Zhejiang, China)52 using samples of serum or plasma.
Automated methodsSeveral techniques for the rapid detection of SARS-CoV-2 by the automated detection of antibodies in less than one hour have recently been introduced. Some of these tests have already been approved by the FDA and they will be awarded the CE mark in the near future.
SARS-CoV-2 IgG Abbott. Chemoluminescent microparticle immunoanalysis (CMIA) detects IgG antibodies against N protein of the nucleocapsid in serum and plasma is a two-stage automated system. In the first phase paramagnetic microparticles coated in SARS-CoV-2 antigen are bound to the IgG antibodies contained in the sample, washed and then a conjugate of human anti-IgG antibodies marked with acridine is added and incubated. Subsequently, after the addition of the preactivating and activating solution the resulting chemoluminescent reaction is read.
LIAISON SARS CoV-2 IgG. This a qualitative assay using chemoluminescent technology (CLIA) that detects IgG antibodies against the S1 and S2 domains of SARS-CoV-2 S protein.
Other pharmaceutical companies are developing new automated assays for the study of IgG antibodies against SARS-CoV-2.
MAGLUMI 2019-nCoV IgG and IgM. This is a quantitative assay using chemoluminescent technology (CLIA) that detects IgG and IgM antibodies against the S protein and N protein of the virus.53
Other assays are available in the market with a monotest presentation that includes all of the necessary reagents, such as COVID-19 VIRCLIA® IgM + IgA MONOTEST, which also detects the presence of IgA.
Semi-automated methodsFew kits are commercialised for the study of IgM and IgG against SARS-CoV-2, and most of them are ELISA (enzyme-linked immunoassay) to be carried out in microplates with 96 wells lined with the S protein of the virus, followed by colorimetric developing (COVID-19 Human IgM IgG Assay Kit Abnova, COVID-19 ELISA IgG e IgM + IgA Vircell).
Epidemiological studies to gain knowledge about the virusStudy of its biological properties: cell cultureSARS-CoV-2 was isolated in cell cultures from the start of the outbreak in China, and this is of enormous interest as it makes it possible to determine whether a certain value of the threshold cycle or Ct obtained by PCR genome amplification methods correlates with virus able to be transmitted between human beings. Positive PCR in a nasopharyngeal sample may last for up to 22 days or longer after the start of symptoms.54,55 Nevertheless, the infective virus (determined by isolation in culture) is only detectable during up to 8 days after the appearance of symptoms in mild cases.55 It has been proven that the minimum viral load necessary to isolate the virus using saliva is 106 copies of RNA/mL. Contagion is estimated to occur from 2−8 days prior to the symptoms and 7 days afterwards.54
The chief applications of the isolation in cell cultures of SARS-CoV-2 include: a) research into antiviral drugs using the experimental capacity of new substances to inhibit the growth of the virus; b) research into the viral pathogenesis by studying transmission chains in human beings, the severity of infection, the viral load deriving from the said infections and the capacity of the virus to infect different organs, and c) research into the stability of the virus, showing its capacity to remain infective on surfaces or under certain environmental conditions, temperature and light (https://www.cdc.gov/coronavirus/2019-ncov/php/grows-virus-cell-culture.html).
Study of its genomic characteristics: mass sequencingGenetic sequencing is a technique used in practically all biological and biomedical research laboratories. The reference technique is Sanger sequencing, which by using fluorescent techniques and capillary electrophoresis makes it possible to automate the sequencing process, it is applied to knowledge of the virus genome, and more specifically to respiratory viruses. Over the years new techniques have arisen that make it possible to obtain millions of sequencing reactions simultaneously and very quickly, using Illumina and IonTorrent technologies, and they have been considered to be part of second generation genome sequencing. The third generation of techniques is composed of methods that are able to sequence DNA molecules without previous amplification, and they have been developed by Pacific BioSciences and Oxford Nanopore.56,57
Sequencing the SARS-CoV-2 genome is an enormous challenge, to know its genome and its variability. Unprecedented sequencing work has been performed from the start of the pandemic to obtain sequences very quickly, so that they can be analysed very soon after detection and diagnosis in patients. The aim of genome analysis is to generate knowledge for research into drugs for treatment and vaccines for prevention. Sequencing data also generate results that make it possible to evaluate diagnostic kits, tracing the origin of the virus, studying the transmission routes and identifying the options for strategic intervention. A protocol has been published in the protocol platform protocols.io (https://www.protocols.io/) for the mass sequencing of SARS-CoV-2 using Oxford Nanopore technology, and this is being updated by its different users (labworm.com/tool/protocolsio).58 Other approaches use Illumina and IonTorrent technologies.
A total of 16,000 genome sequences are now stored in the GISAID database.59 Analysis of the sequences in different countries uses the basic strain which is considered the reference, Wuhan-Hu-1/2019. This was sequenced on 19 January 2020 and deposited with access number MN908947 in the GenBank sequences database, and EPI-ISL-402,119 in the GISAID sequences database. This strain, together with other contemporary ones, is used as the basis for phylogenetic analyses. They are also used as the basis for structural studies of the proteins associated with the new sequences. Spanish sequences have been used to calculate that the evolution of SARS-CoV-2 has a range of 1.08 × 10−3 to 1.87 × 10−3 substitutions per site per year.60
Working of the national epidemiological monitoring network in connection with COVID-19The epidemiological monitoring of transmissible diseases is one of the activities traditionally associated with public health. Such diseases are studied in a department which has the mission of improving our knowledge about them and outbreaks of any type, through monitoring and epidemiological and public health research. The final aim is to supply quality information to guide the control and prevention of transmissible diseases, as well as to guide the design, implementation and evaluation of medical policies.
Monitoring structure is based on the work of the National Epidemiological Monitoring Network (Red Nacional de Vigilancia Epidemiológica) (RENAVE), which is governed by the National Epidemiology Centre of the Instituto de Salud Carlos III (ISCIII). This decentralised structure reflects the regionalised organisation of our country. It is composed of technical public health professionals in local, regional and state health departments. The Network responds to needs for information which arise in health authorities and all of the professionals who, in different fields and with different responsibilities, need to know the symptoms, risk patterns and distribution of transmissible diseases in the population. The RENAVE brings together the monitoring process, including: a) the notification of cases and b) epidemiological research into the said cases of transmissible diseases.
In the current pandemic, the Ministry of Health, through the Health Emergency Warnings and Coordination Centre (CCAES), is in permanent contact with the autonomous communities (AC) through the Warning System and Plans for Preparation and Response. The National Centre for Epidemiology (CNE) also takes part, as does the National Microbiology Centre (CNM) in the ISCIII. Coordination with international bodies (the WHO, the European Centre for Disease Control [ECDC] and the European Commission [EUC]) is basic for the evaluation of risks in situations and coordinating measures to respond.
From the start of the crisis an action protocol was implemented after the appearance of cases in Spain, and this has been updated during 2020. This protocol includes the new coronavirus case Notification Document, the information in which is sent from the AC to the CNE and the CCAES, as well as the indications for sending samples to the CNM.
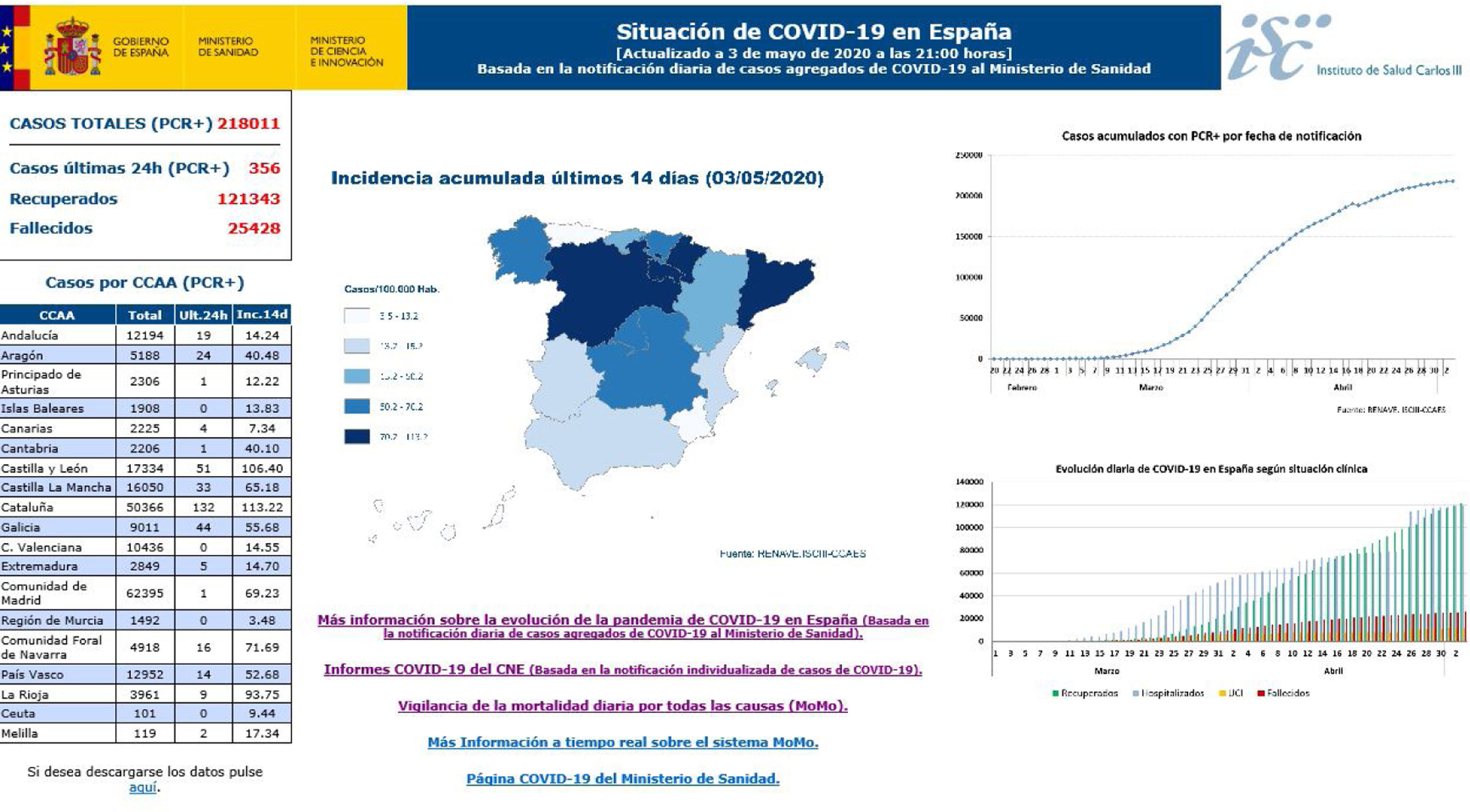
27 reports on the situation were prepared before 30 April, collecting AC data and notifying the Ministry of Health every day about the number of confirmed accumulated cases of COVID-19: the total number of cases, cases in healthcare professions, hospitalisations, admissions to ICU, deaths and cases that had recovered. The AC completed an individualised survey for each one of the said cases, and this includes epidemiological and clinical information agreed and approved by the Warning System and Plans for Preparation and Response and the RENAVE, together with notification using the computerised platform SiViEs (the Monitoring System in Spain), which is managed by the CNE. From the start of the COVID-19 pandemic in Spain cases have been monitored based on the universal reporting of all of the confirmed cases of COVID-19 identified in each AC. The aggregate data referring to COVID-19 (Fig. 2) may be consulted in https://covid19.isciii.es/
It is important to state that the monitoring of excess mortality rates due to all causes takes place using a monitoring system known as MoMo. This records excess mortality due to all causes, evaluating the impact on the mortality of the population of all events that may be a public health threat. Its results may support risk evaluations corresponding to the said events and help to guide a suitable public health response and the development of policies to control such events. These can be consulted in Informes MoMo 2020, at https://cnecovid.isciii.es/
Post mortem microbiological studies with reference to SARS-CoV-2: applicabilityInterest and objectives in the post mortem detection of SARS-CoV-2The main medical and legal circumstances in which it is of interest to detect COVID-19 as a diagnostic technique in forensic autopsy samples are: a) suspected (unconfirmed) cases of COVID-19 in natural deaths b) sudden or unexpected of adults or children in which COVID-19 has to be ruled out as a possible cause of death; c) reports of poor praxis in which infection may be involved in the cause of death; d) violent deaths in which the circumstances make it advisable to determine this factor (e.g., suicide, the possibility of a covert crime), and e) any other situation in which the forensic official considers it to be necessary.
As well as questions of medical and legal importance, the forensic doctor is dealing with a new infectious disease. As such, he is in an optimum position to:
- none-
Identify the physiopathology and mechanism of action of this new infectious agent.
- none-
Identify patterns of infection in different risk populations and age groups.
- none-
Contribute to the epidemiological study of the pandemic.
- none-
Optimise post mortem diagnostic techniques.
To date, practical experience of the post mortem detection of COVID-19 infection has been restricted by the small number of autopsies performed on these patients in the affected countries.24 The few published papers on the results of autopsies in cases of COVID-19 are restricted to certain organs and/or tissues, or they use minimally invasive techniques, and complete autopsies are only described in a few cases.20,24,25,61,62 With some exceptions, the majority of these publications do not offer many details on the microbiological studies that were performed. In the majority of these studies the detection of the pathogen was performed ante mortem with the usual samples taken in clinical practice (NPS or pharyngeal swab), or the samples used for diagnosis or when they were taken is not specified. In the publications that state that microbiological diagnosis took place post mortem, the technique used was PCR.21,63,64 In these cases, the sample analysed was a pharyngeal swab,64 and only a few cases used samples taken from the lower airways, such as lung parenchyma swabs62 or lung necropsies fixed in paraffin.21 Only exceptionally was electron microscopy used to verify the presence of viral inclusion particles in the lungs20 or kidneys.61
No study was found in the bibliography consulted which made parallel use of more than one type of sample for microbiological analysis, so that it is impossible to make any recommendation on this basis as to which sample may be the most recommendable to use in corpses.
Recommendations for the determination of COVID-19 in corpsesMolecular techniquesForensic protocols in Spain for the study of COVID-1965 indicate that samples should be taken prior to autopsy, more specifically NPS and pharyngeal swab. These would be the most suitable samples for the purpose of screening for COVID-19 to decide whether to carry out an autopsy, depending on whether the result is negative or positive, respectively. These samples must be collected in a virus transport medium or an inactivation medium; the latter, as well as guaranteeing the stability of the virus, makes it possible to inactivate for increased biosafety during transport and the start of analysis. These samples will be used for molecular analysis.
In any case, if it is decided to perform a complete, directed or minimally invasive autopsy, other additional samples can be taken from the lower airways for microbiological diagnosis. These samples can be bronchial swabs or swabs of the pulmonary parenchyma, and even a small fresh lung biopsy. These samples may be highly useful when the virus is undetectable in the nasopharynx – which may occur in certain phases of the disease – but is detectable in the lower airways, as is sometimes the case with the influenza virus or other respiratory viruses. Additionally, unpublished data from post mortem studies in COVID-19 (personal communication by Nihan Ziyade and Neval Elgormus) show that the tracheal swabs may be positive when the NPS is negative. All of these must be taken into account, especially in cases with a suspicion of COVID-19 and a negative result in the NPS or pharyngeal swabs.
Another sample that may be of interest is a rectal swab, as excretion of the virus in stools is more prolonged (up to 30 days), although it has to be taken into consideration in the interpretation of these results that the finding does not necessarily indicate that the dead person was in the acute phase of the disease, but rather than it is a sign of an infection in the past.
As was pointed out above, the interpretation of PCR in corpses must be approached with care. If a NPS result is clearly positive, in a pharyngeal or lower airway sample, it will be assumed that the dead individual had COVID-19 infection. Nevertheless, this does not always imply that the patient is in the acute phase of the disease, given that series of patients have been published in which the viral RNA of this pathogen persists in clinical samples without the virus being viable.
On the contrary, if a result is negative, this negativity may not be due to the absence of infection, but rather to other reasons, of which the main ones are:
- none-
The sample was not taken properly.
- none-
The virus is no longer detectable in the pharynx or nasopharynx, but it may be in the lower airways because of the evolution of the disease.
- none-
The infection is remitting and the viral load, as it is very low, is below the detection limit of the technique.
There are currently no well-founded data on which to base the use of the so-called fast serological tests for screening and to recommend their routine use in corpses, given that this may lead to errors, according to the above considerations.
Respecting the use of automated or semi-automated classical serological tests, it would make sense to use these in the following circumstances:
- none-
Deaths with a negative PCR result and the suspicion of COVID-19 infection, i.e., to confirm the infection in the absence of a positive PCR. In these cases the evolution of the infection must have lasted for longer than 7 days.
- none-
When a “past infection” has to be confirmed by IgG determination, in connection or not with the negative result of an ante-mortem PCR test.
Due to these reasons the forensic protocols at national level have included serum sampling, with the aim of performing serological studies. Moreover, these protocols have also included taking samples of peripheral blood with EDTA, to detect a possible viremia. However, the diagnostic utility of this would be very limited, as there are no studies which prove the presence of the virus in the blood or urine.66
In any case, the final interpretation of the results obtained must always take the clinical situation of the dead individual into account, so that it would be advisable to have data from their clinical history, if this is available, as well as the interview with those who they lived with, all with the aim of tracing the start of the symptoms, if there were any. We must not forget that, above all in elderly patients, some symptoms may be confused with manifestations of their age (chronic cough, comorbidity or cognitive deterioration, etc.).
Other microbiological studies of interest in those who died due to COVID-19Although from a forensic point of view deaths due to COVID-19 have easily distinguishable characteristics, the proven fact that these patients may have suffered different types of coinfection at the moment of death makes it necessary to perform complementary microbiological tests to clarify this possibility. If the corpse is not opened, then logically the possibility of detecting these coinfections will be reduced. It is always possible to take additional NPS and pharyngeal swabs to use for bacterial culture and molecular analysis for respiratory viruses. Nevertheless, if these tests are negative, it does not mean that there is no infection of the lower airways. It is therefore advisable to also take a bronchial swab and lung biopsy, in the case of a traditional autopsy,67 or syringe aspirate of the lung if a minimally invasive autopsy is performed, following ESGFOR recommendations (the European Study Group of Forensic and Post-mortem Microbiology of the ESCMID, the European Society of Clinical Microbiology and Infectious Diseases).68
As in any other post mortem study, the most frequent analyses are bacterial and fungal culture techniques, antigen detection techniques and molecular techniques.69
To detect the bacteria that may appear in COVID-19 coinfections bacterial cultures of samples may be made in conventional solid and liquid media, under similar incubation conditions and times to those used for the studies of living patients. Atypical bacteria (Legionella sp. and C. pneumoniae) may be analysed using antigen techniques (enzyme immunoanalysis or inmunochromatography) or molecular techniques. When there is the suspicion of a fungal infection, samples must be inoculated in Sabouraud agar with antibiotic, although it is true that Aspergillus grows well in almost all culture media, for bacteria as well as for fungi. Fungal antigens such as galactomannan are useful to detect the presence of this fungus, as it most especially a search for its DNA. The detection of nucleic acids using molecular techniques is useful to detect any of the microorganisms that may accompany SARS-CoV-2, although virological studies are especially relevant here.
All molecular studies commence with the extraction of nucleic acids from a sample. This is usually done using automatic platforms that guarantee that the material obtained is of suitable quality. This material is subjected to different types of PCR that may be combined with other techniques such as microarrays. To detect the presence of respiratory viruses other than SARS-CoV-2 it is highly useful to use commercial microarrays that make it possible to analyse the 17 most common respiratory viruses in humans (Clart Pneumovir. Genómica S.A.U.). Other techniques, such as the Luminex NxTAG or the Biomerieux Filmarray (multiple PCRs), make it possible to easily and simultaneously detect the main respiratory viruses as well as the bacterial which cause atypical pneumonias, and even to quantify them in some cases.
Conflict of interestsThe authors have no conflict of interests to declare.
The authors would like to express their thanks to the members of the ESGFOR Group (European Study Group of Forensic and Post-mortem Microbiology, of the ESCMID, the European Society of Clinical Microbiology and Infectious Diseases) for their help and experience regarding the COVID-19 pandemic, most especially Dr. Nihan Ziyade and Dr. Neval Elgormus (Ministry of Justice, Council of Forensic Medicine, Post-mortem Microbiology Laboratory, Istanbul, Turkey).
Please cite this article as: Fernández-Rodríguez A, Casas I, Culebras E, Morilla E, Cohen M C, Alberola J, COVID-19 y estudios microbiológicos post mortem. Rev Esp Med Legal. 2020;46:127–138.