Insertion–deletions for human identification (HID-INDELs) allow solving peculiar forensic situations when autosomal STRs are insufficient. Although limitations were predicted since the forensic implementation of biallelic markers, formal evaluation of these restrictions is scarce. Particularly, to define the informativity provided by HID-INDELs in kinship analysis is useful to avoid wasting work, resources, and –finally– disappointments.
Material and methodsFor this reason, we analyzed the 38-plex HID-INDEL system in 25 Mexican families including father, daughter, and mother, whose kinship was previously established with 22 autosomal STRs.
Results and discussionFrom genotypes of unrelated individuals, we updated allele frequencies and forensic parameters of the Jalisco state (West, Mexico), by increasing the population sample size from 62 to 112. Among the forensic a priori parameters, the Typical paternity index (PI) of the 38plex HID-INDEL system showed important differences regarding the PI and probability of paternity (W) estimated herein from real paternity cases, generally undervaluing the observed informativity of these 38-plex HID-INDEL system. Conversely, the studied HID-INDEL loci offered confident kinship conclusions based on the paternity index (PI ≥10,000) and probability of paternity (W ≥ 99.99%) in 68% of the standard trio cases (18/25), and only 12% of duo paternity cases (6/50) (motherless and fatherless). In fact, 14% of duo paternity cases (7/50) did not reach minimum requirements to stablish paternity (IP < 100; W < 99%).
ConclusionsWe updated a Mexican population database for 38 HID-INDEL loci, and we described their proficiency from real paternity cases, detailing some limitations non-previously specified.
Las inserciones-deleciones para identificación humana (HID-INDEL), permiten resolver situaciones forenses peculiares cuando los STR autosómicos son insuficientes. Aunque se predijeron sus limitaciones desde la implementación forense de marcadores bialélicos, la evaluación formal de estas restricciones es escasa. Particularmente es útil definir su informatividad en el análisis de parentesco, para evitar desperdiciar trabajo, recursos y –finalmente– decepciones.
Material y MétodosPor este motivo, analizamos el sistema de 38plex HID-INDELs en 25 familias mexicanas entre padre, hija y madre, cuyo parentesco se estableció previamente con 22 STR autosómicos. A partir de los individuos no emparentados, actualizamos las frecuencias alélicas y los parámetros forenses del estado de Jalisco (Oeste, México), aumentando el tamaño de la muestra poblacional de 62 a 112.
Resultados y discusiónEntre los parámetros forenses a priori, el índice de paternidad típico (IPT) del sistema 38plex HID-INDEL mostró diferencias importantes con respecto al IP y la probabilidad de paternidad (W) derivados de casos reales de paternidad, generalmente subestimando la informatividad observada de ese sistema. Sin embargo, los 38 HID-INDELs ofrecieron conclusiones de parentesco confiables basadas en el índice de paternidad (IP ≥ 10,000) y la probabilidad de paternidad (W ≥ 99,99%) sólo en el 68% de los casos tríos estándar (18/25), y el 12% de casos de paternidad dúo (6/50) (sin madre y sin padre). De hecho, el 14% de los casos de paternidad dúo (7/50) no alcanzó los requisitos mínimos para establecer la paternidad (IP <100; W < 99%).
ConclusionesEn este trabajo actualizamos una base de datos de población mexicana para 38 HID-INDELs y describimos su eficiencia en casos reales de paternidad, detallando algunas limitaciones no especificadas previamente.
During the last decades, autosomal Short tandem repeat (STR) loci have constituted the markers of choice for Human identification (HID) purposes.1 However, poor-quality samples commonly found in forensic casework generate partial STR profiles that complicate reaching conclusions (e.g. ancient or degraded DNA).2 Similarly, the relatively high mutation rate of STRs generates sporadic inconsistencies in kinship analysis.3 Although Single nucleotide polymorphisms (SNPs) have been implemented for HID purposes, they require instruments non-available in the majority of forensic genetic labs and/or involve no conventional methodologies.2,4 Conversely, Insertion–Deletion length polymorphisms (INDELs) have been easily incorporated in forensic casework by means of short amplicon multiplexes followed by traditional capillary electrophoresis.5–7 Particularly, both SNPs and INDELs biallelic loci have been useful for analysis of highly degraded skeletal remains,8 and to deal with occasional STR mutations in paternity testing due to their low mutation rate.9–11 Consequently, genetic systems based on HID-INDELs have been studied in worldwide populations to estimate allele frequencies and a priori forensic parameters, such as heterozygosity, power of discrimination (PD), power of exclusion (PE), and typical paternity index (IP), principally.12–14 However, informativity weaknesses due to their biallelic condition have been described for kinship analysis, particularly in motherless duo-cases,15 and when the alleged father is a close relative of the real one.9,10,16 Interestingly, some parameters a posteriori such as the Paternity index (PI)17 have demonstrated to be useful to evaluate the actual contribution of genetic markers in conventional kinship analyses,18 which would be worthy to avoid wasting work, resources, and –finally– disappointments.
For this reason, we analyzed the 38-plex INDEL system in 25 Mexican families from the Jalisco state, including the father (F), mother (M), and daughter (D) with previously established kinship based on 22 autosomal STRs. The aims of this study were the following: 1) to evaluate the proficiency provided by the 38plex INDEL system to confirm kinship (i.e. PI) in these families fitted as standard trio and duo paternity cases (motherless and fatherless); 2) to update the forensic parameters of the studied Mexican population using INDEL genotypes from unrelated individuals and previously reported.13
Materials and methodsStudied sampleA total of 75 volunteers from 25 Mexican families from the Jalisco state (West, Mexico) were analyzed. Genetic relationships were previously confirmed with 22 autosomal STRs by the Powerplex Fusion kit (Powerplex Fusion System, Promega Corp.) displaying a PI ≥1.46E+12. These 25 families also constitute 25 standard paternity cases (trio), with 50 unrelated persons (fathers plus mothers), plus 25 individuals corresponding to the offspring. All individuals of each family signed a written informed consent agreeing to the ethical guidelines of the Helsinki Declaration. This study was approved by the local Ethical Research Committee of the Research Institute in Molecular Genetics.
Laboratory analysisGenomic DNA was collected from bloodstain samples impregnated on FTA papers. Bloodstain punches of 1.2 mm washed with FTA Purification Reagent (Whatman™) and DNAsa-free water, were directly used as DNA samples for further analysis. Genotypes for the 38 autosomal INDELS were obtained using the PCR multiplex protocol previously described for this genetic system.5 Separation and detection of PCR products were performed with an ABI Prism 3130 Genetic Analyzer, and using the GeneMapper v3.2 software (Applied Biosystems, Foster City, CA).
Data analysisFirstly, for each Mexican family, the paternity index (PI) was estimated by INDEL locus and combined PI for each standard trio case (n = 25). Moreover, PI's were computed omitting one parent, adapting data as motherless and fatherless paternity duo-cases (n = 50). When the probability of paternity (W) was estimated, a flat a priori probability of 0.5 was taken into account for both paternity hypotheses: kinship and unrelated. With the INDEL genotypes of unrelated individuals, specifically fathers and mothers (n = 50), plus those previously reported (n = 62),13 we were able to updated the forensic parameters of the Jalisco state (n = 112). The updated allele frequencies were used to compute the PI values with PATPCR, a free Excel spreadsheet designed by JA Luque (Barcelona, Spain, 2002). Although individuals of the paternity cases are included in the updated Mexican population database, a minimal effect is expected given the biallelic condition of INDEL markers. The Microsoft Excel spreadsheet Powerstats19 was used to compute the following a priori forensic parameters: allele frequencies, observed heterozygosity (Ho), expected heterozygosity (He), power of discrimination (PD), typical paternity index (TPI), and polymorphic information content (PIC). However, the power of exclusion (PE) was computed with an specific formula for biallelic loci [PE = pq (1-pq)]. Finally, exact tests to assess Hardy–Weinberg expectations (HWE) and linkage disequilibrium (LD) between pairs of loci were performed using the GDA software.20
Results and discussionsKinship informativity of the 38plex INDEL systemThe complete 38-plex INDEL genotypes of the Mexican families are available in the Supplementary table S1. As expected by the low mutation rate of the INDEL loci,9–11 any exclusion parent-daughter was observed, and PIs were estimated to evaluate the performance of the 38-plex INDEL system for kinship analysis (Supplementary table S2a and S2b). In duo paternity cases, the average PI values in fatherless (32,119) and motherless cases (40,716) were similar (P = 0.8579; t-student test), as could be expected. However, differences between PIs from trio and duo paternity cases were evident (P < 0.0001; t-student test), as shown in Fig. 1. Concerning the informativity by INDEL locus, R09 was the most informative locus in trio paternity cases with the largest range (average PI = 2.302; range = 0.5803–7.2254) (Table 1), which is associated with the extreme allele frequencies of R09 –closer to zero and one– that result in the largest and smallest PI values, respectively (Table 1). Based on duo paternity cases, R06 was the most informative INDEL locus (average PI = 1.5067). Conversely, although R03 displayed the minimum average PI (1.1407), R09 showed the smallest PI value (0.5803) (Table 1).
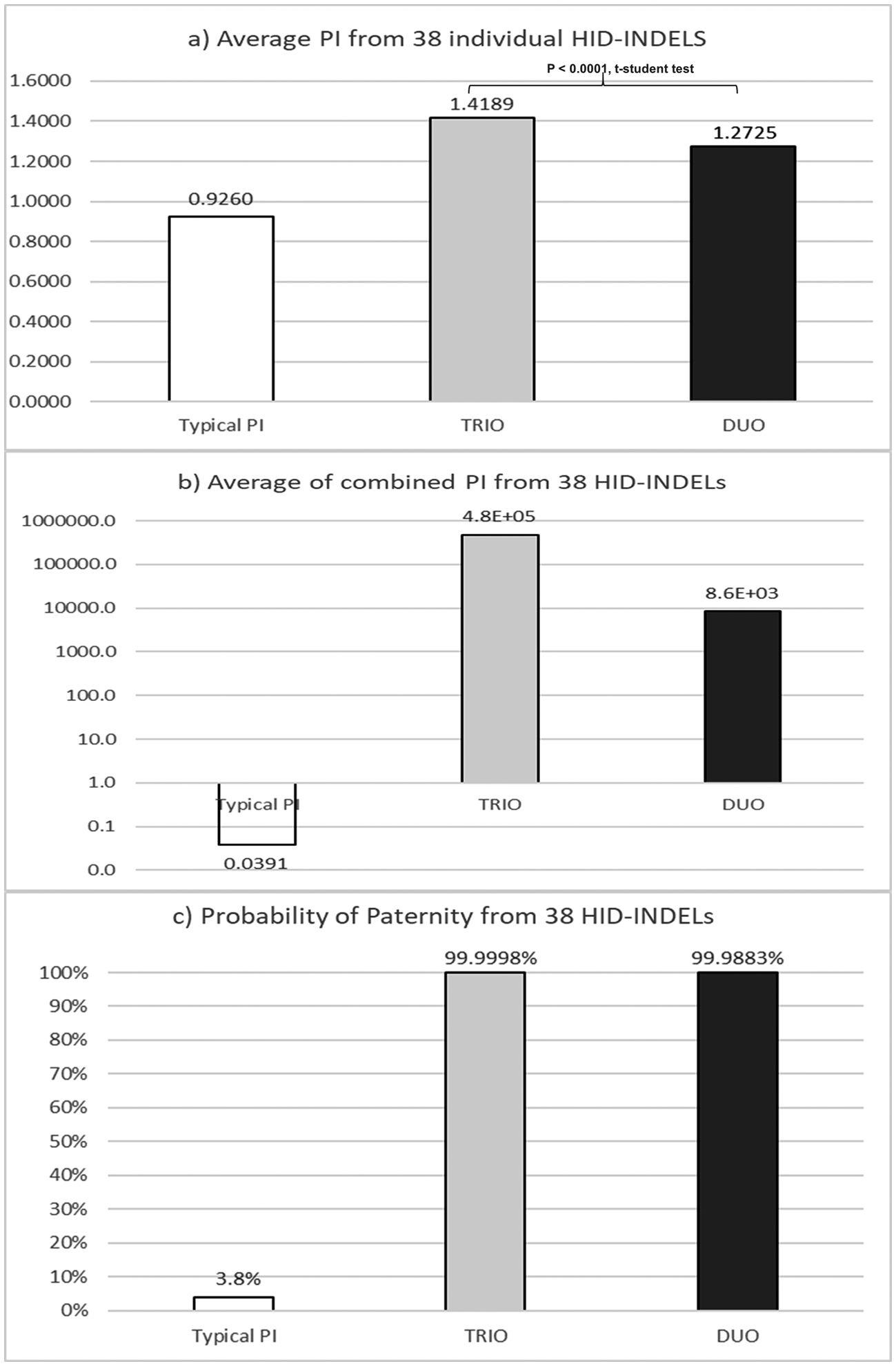
Comparison of kinship results form typical paternity index (TPI) versus those from standard trio an duo paternity cases for the 38 HID-INDELs5: a) Average PI of individual markers; b) Combined from average PI of markes; c) Probability of paternity (W) from values described in incise b).
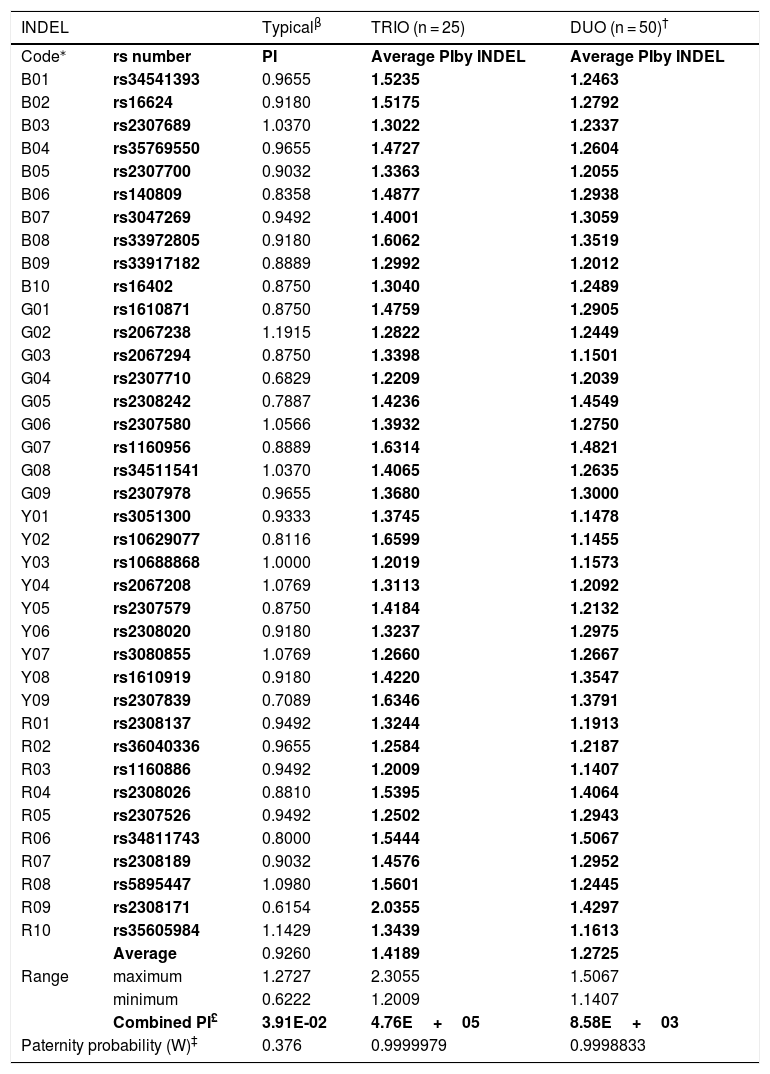
Comparison of the typical paternity index (TPI) and average PI from standard trio and duo paternity cases based on 25 families from a Mexican population.
| INDEL | Typicalβ | TRIO (n = 25) | DUO (n = 50)† | |
|---|---|---|---|---|
| Code⁎ | rs number | PI | Average PIby INDEL | Average PIby INDEL |
| B01 | rs34541393 | 0.9655 | 1.5235 | 1.2463 |
| B02 | rs16624 | 0.9180 | 1.5175 | 1.2792 |
| B03 | rs2307689 | 1.0370 | 1.3022 | 1.2337 |
| B04 | rs35769550 | 0.9655 | 1.4727 | 1.2604 |
| B05 | rs2307700 | 0.9032 | 1.3363 | 1.2055 |
| B06 | rs140809 | 0.8358 | 1.4877 | 1.2938 |
| B07 | rs3047269 | 0.9492 | 1.4001 | 1.3059 |
| B08 | rs33972805 | 0.9180 | 1.6062 | 1.3519 |
| B09 | rs33917182 | 0.8889 | 1.2992 | 1.2012 |
| B10 | rs16402 | 0.8750 | 1.3040 | 1.2489 |
| G01 | rs1610871 | 0.8750 | 1.4759 | 1.2905 |
| G02 | rs2067238 | 1.1915 | 1.2822 | 1.2449 |
| G03 | rs2067294 | 0.8750 | 1.3398 | 1.1501 |
| G04 | rs2307710 | 0.6829 | 1.2209 | 1.2039 |
| G05 | rs2308242 | 0.7887 | 1.4236 | 1.4549 |
| G06 | rs2307580 | 1.0566 | 1.3932 | 1.2750 |
| G07 | rs1160956 | 0.8889 | 1.6314 | 1.4821 |
| G08 | rs34511541 | 1.0370 | 1.4065 | 1.2635 |
| G09 | rs2307978 | 0.9655 | 1.3680 | 1.3000 |
| Y01 | rs3051300 | 0.9333 | 1.3745 | 1.1478 |
| Y02 | rs10629077 | 0.8116 | 1.6599 | 1.1455 |
| Y03 | rs10688868 | 1.0000 | 1.2019 | 1.1573 |
| Y04 | rs2067208 | 1.0769 | 1.3113 | 1.2092 |
| Y05 | rs2307579 | 0.8750 | 1.4184 | 1.2132 |
| Y06 | rs2308020 | 0.9180 | 1.3237 | 1.2975 |
| Y07 | rs3080855 | 1.0769 | 1.2660 | 1.2667 |
| Y08 | rs1610919 | 0.9180 | 1.4220 | 1.3547 |
| Y09 | rs2307839 | 0.7089 | 1.6346 | 1.3791 |
| R01 | rs2308137 | 0.9492 | 1.3244 | 1.1913 |
| R02 | rs36040336 | 0.9655 | 1.2584 | 1.2187 |
| R03 | rs1160886 | 0.9492 | 1.2009 | 1.1407 |
| R04 | rs2308026 | 0.8810 | 1.5395 | 1.4064 |
| R05 | rs2307526 | 0.9492 | 1.2502 | 1.2943 |
| R06 | rs34811743 | 0.8000 | 1.5444 | 1.5067 |
| R07 | rs2308189 | 0.9032 | 1.4576 | 1.2952 |
| R08 | rs5895447 | 1.0980 | 1.5601 | 1.2445 |
| R09 | rs2308171 | 0.6154 | 2.0355 | 1.4297 |
| R10 | rs35605984 | 1.1429 | 1.3439 | 1.1613 |
| Average | 0.9260 | 1.4189 | 1.2725 | |
| Range | maximum | 1.2727 | 2.3055 | 1.5067 |
| minimum | 0.6222 | 1.2009 | 1.1407 | |
| Combined PI£ | 3.91E-02 | 4.76E+05 | 8.58E+03 | |
| Paternity probability (W)‡ | 0.376 | 0.9999979 | 0.9998833 | |
The color code of the INDEL columns are related with the fluorocroms used during the PCR and detected in the capillary electrophoresis, and is in agreement with the original description.5
The generally accepted minimum standard PI for inclusion of paternity is ≥100,21 but presently the majority of the labs attached to the English-Speaking Group of the International Society of Forensic Genetics (ESG-ISFG) require a PI≥10,000,22 which is probably required in most worldwide paternity testing labs. These PIs correspond to probabilities of paternity (W) of 99 and 99.99%, respectively, assuming a flat a priori probability (p = 0.5). From the studied Mexican families in standard paternity trio-cases, the 38-plex INDEL system displayed an average combined PI = 1.63E+05 (range: 1.09E+02 to 1.29E+06). However, 32% (8/25) of these PI values are below the limit required in most of the paternity laboratories (PI<10,000)22 (Supplementary table S2a).
On the other hand, the Mexican families adapted as paternity duo-cases showed an average combined PI = 3.64E+04 (range: 5.98 to 9.55E+05), but only 12% of these values (6/50) reached the required limit to establish paternity (PI>10,000) (Supplementary table S2b). Even 14% (7/50) of these paternity duo-cases neither reached the minimum standard for inclusion of paternity (PI<100) 21 (Supplementary table S2b). In brief, the estimated performance of the 38-plex INDEL system to establish confident kinship from real standard trio and duo cases was only 68% and 12%, respectively.
These results highlight the role of the 38-plex system as a complementary genetic system to establish paternity. This is in agreement with the limitations described for biallelic markers to solve duo paternity cases,15 and when the alleged father (AF) is a close relative of the real father.9,10 For instance, to discard the four-step mutation hypothesis between the AF and child in the autosomal STR D22S1045, concerning the brother of the AF paternity hypothesis.16 In this study, although the AF paternity hypothesis was favored with the 81 HID markers analyzed (LR = 13.4; W = 93.1%), 58 INDELs –by themselves– were not conclusive enough (LR = 110.3, W = 99.1%). Conversely, the 38-plex INDEL system has contributed successfully to solve complex paternity cases, such as supporting the hypothesis of maternal uniparental disomy of chromosome 2, despite the exclusions presence.23
The a priori forensic parameter for standard paternity cases, known as Typical paternity index (TPI),24 was compared with its corresponding a posteriori analog based on the Mexican paternity cases studied herein (Fig. 1). By INDEL locus, the average TPI was minor than one (PI = 0.94; range: 0.6222 to 1.2727) (Table 1). This finding means that –theoretically– each INDEL marker commonly does not contribute statistically to confirm true paternities, despite the observed parent–child match (Fig. 1A). Conversely, the average PI estimated herein was larger than one, for both standard trio (PI = 1.4189; range = 1.2009–2.3055) and duo paternity cases (PI = 1.2725; range = 1.1407–1.5067) (Table 1). Consequently, INDELs contribute –modestly– to establish previously verified kinship (Fig. 1A). Interestingly, the product of the average TPI from the 38 INDELs did not support the paternity hypothesis (TPI = 0.0688; W = 6.43%) (Table 1; Fig. 1B, C). On the other hand, the product of the average PI from the 38 INDELs helped to verify the kinship in both trios (PI = 4.76E+05; W = 99.99979%) and duo paternity cases (PI = 8.58E+03; W = 99.98833%) (Fig. 1B, C). In brief, these results show that TPI undervalues the informativity provided by HID-INDELs –and consequently SNPs– to verify kinship; therefore, this a priori forensic parameter should be redesigned according to the low polymorphism of these biallelic markers.
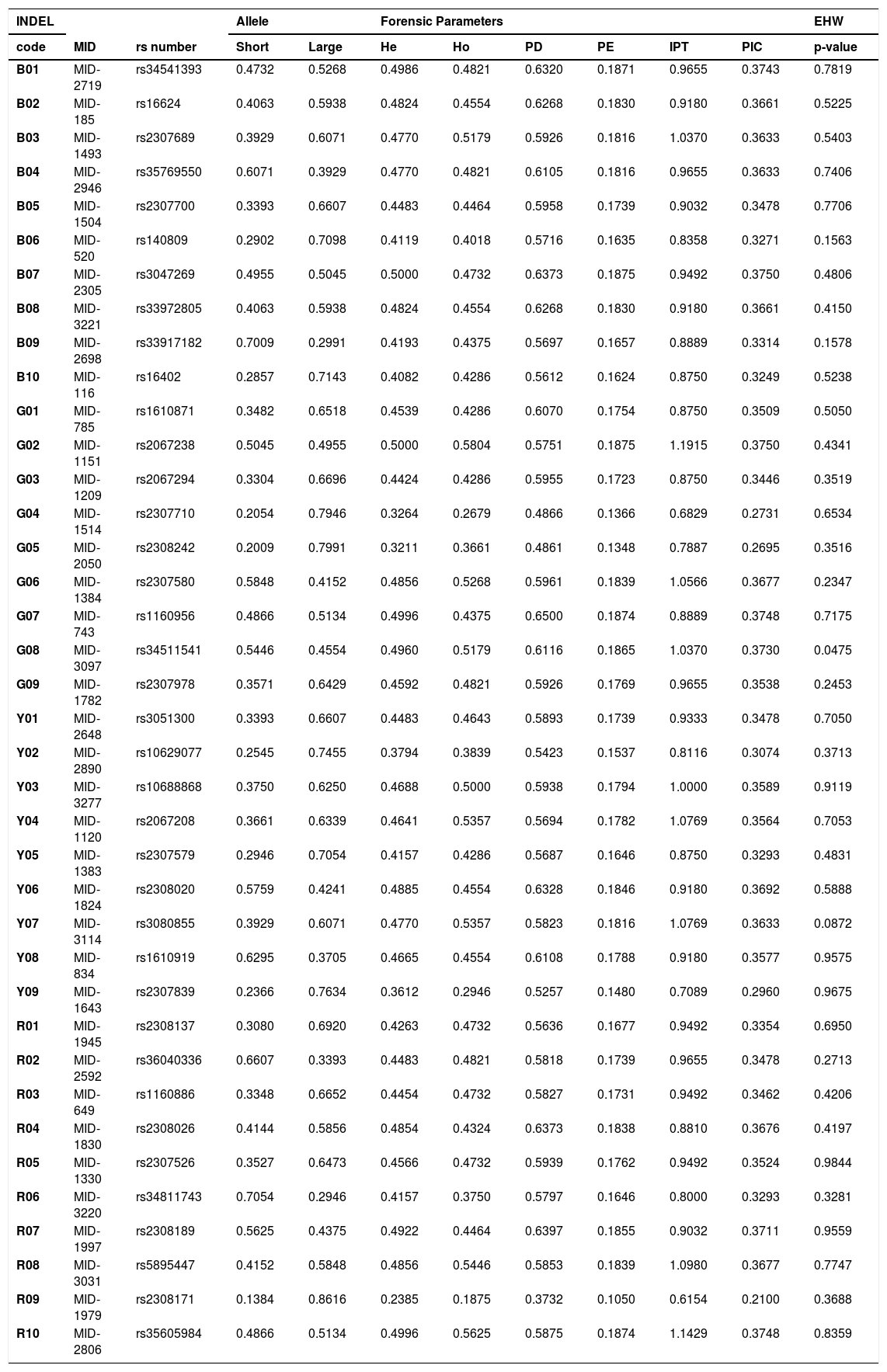
Updating forensic parameters in a Mexican populationThe INDEL genotypes from unrelated individuals studied herein (father and mother), were combined to the corresponding previously reported Mexican population dataset (Supplementary table S3).13 Therefore, we updated the allele frequencies and forensic parameters of the 38 HID-Indels in the Jalisco state (Table 2), increasing the sample size from 62 to 112 individuals, improving the parameter's confidence. The average heterozygosities observed (Ho) and expected (He) in these 38 INDELs were 0.4505 and 0.4461, respectively. By INDEL, the observed heterozygosity (Ho) ranged from 0.1875 to 0.5804, whereas the expected heterozygosity (He) ranged from 0.2385 to 0.500, for the INDEL loci R09 and G02, respectively. Particularly, R09 showed the lowest of PD, PE, TPI, and PIC values (37.3%, 10.5%, 0.6154 and 21%, respectively). Conversely, G07 showed the highest PD (65%), and G02 showed the highest PE, PIC, and TPI values (18.75%, 37.5%, and 1.19, respectively) (Table 2). Although the combined PD for the 38 INDELS in the Mexican population of Jalisco is large (99.9999999999997%), the combined PE is relatively low (99.93%) regarding what is provided by conventional autosomal STR kits in the same Mexican population.25
Allele frequencies and forensic parameters for 38 HID-INDELs in an updated Mexican sample from the Jalisco state (n = 112).
| INDEL | Allele | Forensic Parameters | EHW | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| code | MID | rs number | Short | Large | He | Ho | PD | PE | IPT | PIC | p-value |
| B01 | MID-2719 | rs34541393 | 0.4732 | 0.5268 | 0.4986 | 0.4821 | 0.6320 | 0.1871 | 0.9655 | 0.3743 | 0.7819 |
| B02 | MID-185 | rs16624 | 0.4063 | 0.5938 | 0.4824 | 0.4554 | 0.6268 | 0.1830 | 0.9180 | 0.3661 | 0.5225 |
| B03 | MID-1493 | rs2307689 | 0.3929 | 0.6071 | 0.4770 | 0.5179 | 0.5926 | 0.1816 | 1.0370 | 0.3633 | 0.5403 |
| B04 | MID-2946 | rs35769550 | 0.6071 | 0.3929 | 0.4770 | 0.4821 | 0.6105 | 0.1816 | 0.9655 | 0.3633 | 0.7406 |
| B05 | MID-1504 | rs2307700 | 0.3393 | 0.6607 | 0.4483 | 0.4464 | 0.5958 | 0.1739 | 0.9032 | 0.3478 | 0.7706 |
| B06 | MID-520 | rs140809 | 0.2902 | 0.7098 | 0.4119 | 0.4018 | 0.5716 | 0.1635 | 0.8358 | 0.3271 | 0.1563 |
| B07 | MID-2305 | rs3047269 | 0.4955 | 0.5045 | 0.5000 | 0.4732 | 0.6373 | 0.1875 | 0.9492 | 0.3750 | 0.4806 |
| B08 | MID-3221 | rs33972805 | 0.4063 | 0.5938 | 0.4824 | 0.4554 | 0.6268 | 0.1830 | 0.9180 | 0.3661 | 0.4150 |
| B09 | MID-2698 | rs33917182 | 0.7009 | 0.2991 | 0.4193 | 0.4375 | 0.5697 | 0.1657 | 0.8889 | 0.3314 | 0.1578 |
| B10 | MID-116 | rs16402 | 0.2857 | 0.7143 | 0.4082 | 0.4286 | 0.5612 | 0.1624 | 0.8750 | 0.3249 | 0.5238 |
| G01 | MID-785 | rs1610871 | 0.3482 | 0.6518 | 0.4539 | 0.4286 | 0.6070 | 0.1754 | 0.8750 | 0.3509 | 0.5050 |
| G02 | MID-1151 | rs2067238 | 0.5045 | 0.4955 | 0.5000 | 0.5804 | 0.5751 | 0.1875 | 1.1915 | 0.3750 | 0.4341 |
| G03 | MID-1209 | rs2067294 | 0.3304 | 0.6696 | 0.4424 | 0.4286 | 0.5955 | 0.1723 | 0.8750 | 0.3446 | 0.3519 |
| G04 | MID-1514 | rs2307710 | 0.2054 | 0.7946 | 0.3264 | 0.2679 | 0.4866 | 0.1366 | 0.6829 | 0.2731 | 0.6534 |
| G05 | MID-2050 | rs2308242 | 0.2009 | 0.7991 | 0.3211 | 0.3661 | 0.4861 | 0.1348 | 0.7887 | 0.2695 | 0.3516 |
| G06 | MID-1384 | rs2307580 | 0.5848 | 0.4152 | 0.4856 | 0.5268 | 0.5961 | 0.1839 | 1.0566 | 0.3677 | 0.2347 |
| G07 | MID-743 | rs1160956 | 0.4866 | 0.5134 | 0.4996 | 0.4375 | 0.6500 | 0.1874 | 0.8889 | 0.3748 | 0.7175 |
| G08 | MID-3097 | rs34511541 | 0.5446 | 0.4554 | 0.4960 | 0.5179 | 0.6116 | 0.1865 | 1.0370 | 0.3730 | 0.0475 |
| G09 | MID-1782 | rs2307978 | 0.3571 | 0.6429 | 0.4592 | 0.4821 | 0.5926 | 0.1769 | 0.9655 | 0.3538 | 0.2453 |
| Y01 | MID-2648 | rs3051300 | 0.3393 | 0.6607 | 0.4483 | 0.4643 | 0.5893 | 0.1739 | 0.9333 | 0.3478 | 0.7050 |
| Y02 | MID-2890 | rs10629077 | 0.2545 | 0.7455 | 0.3794 | 0.3839 | 0.5423 | 0.1537 | 0.8116 | 0.3074 | 0.3713 |
| Y03 | MID-3277 | rs10688868 | 0.3750 | 0.6250 | 0.4688 | 0.5000 | 0.5938 | 0.1794 | 1.0000 | 0.3589 | 0.9119 |
| Y04 | MID-1120 | rs2067208 | 0.3661 | 0.6339 | 0.4641 | 0.5357 | 0.5694 | 0.1782 | 1.0769 | 0.3564 | 0.7053 |
| Y05 | MID-1383 | rs2307579 | 0.2946 | 0.7054 | 0.4157 | 0.4286 | 0.5687 | 0.1646 | 0.8750 | 0.3293 | 0.4831 |
| Y06 | MID-1824 | rs2308020 | 0.5759 | 0.4241 | 0.4885 | 0.4554 | 0.6328 | 0.1846 | 0.9180 | 0.3692 | 0.5888 |
| Y07 | MID-3114 | rs3080855 | 0.3929 | 0.6071 | 0.4770 | 0.5357 | 0.5823 | 0.1816 | 1.0769 | 0.3633 | 0.0872 |
| Y08 | MID-834 | rs1610919 | 0.6295 | 0.3705 | 0.4665 | 0.4554 | 0.6108 | 0.1788 | 0.9180 | 0.3577 | 0.9575 |
| Y09 | MID-1643 | rs2307839 | 0.2366 | 0.7634 | 0.3612 | 0.2946 | 0.5257 | 0.1480 | 0.7089 | 0.2960 | 0.9675 |
| R01 | MID-1945 | rs2308137 | 0.3080 | 0.6920 | 0.4263 | 0.4732 | 0.5636 | 0.1677 | 0.9492 | 0.3354 | 0.6950 |
| R02 | MID-2592 | rs36040336 | 0.6607 | 0.3393 | 0.4483 | 0.4821 | 0.5818 | 0.1739 | 0.9655 | 0.3478 | 0.2713 |
| R03 | MID-649 | rs1160886 | 0.3348 | 0.6652 | 0.4454 | 0.4732 | 0.5827 | 0.1731 | 0.9492 | 0.3462 | 0.4206 |
| R04 | MID-1830 | rs2308026 | 0.4144 | 0.5856 | 0.4854 | 0.4324 | 0.6373 | 0.1838 | 0.8810 | 0.3676 | 0.4197 |
| R05 | MID-1330 | rs2307526 | 0.3527 | 0.6473 | 0.4566 | 0.4732 | 0.5939 | 0.1762 | 0.9492 | 0.3524 | 0.9844 |
| R06 | MID-3220 | rs34811743 | 0.7054 | 0.2946 | 0.4157 | 0.3750 | 0.5797 | 0.1646 | 0.8000 | 0.3293 | 0.3281 |
| R07 | MID-1997 | rs2308189 | 0.5625 | 0.4375 | 0.4922 | 0.4464 | 0.6397 | 0.1855 | 0.9032 | 0.3711 | 0.9559 |
| R08 | MID-3031 | rs5895447 | 0.4152 | 0.5848 | 0.4856 | 0.5446 | 0.5853 | 0.1839 | 1.0980 | 0.3677 | 0.7747 |
| R09 | MID-1979 | rs2308171 | 0.1384 | 0.8616 | 0.2385 | 0.1875 | 0.3732 | 0.1050 | 0.6154 | 0.2100 | 0.3688 |
| R10 | MID-2806 | rs35605984 | 0.4866 | 0.5134 | 0.4996 | 0.5625 | 0.5875 | 0.1874 | 1.1429 | 0.3748 | 0.8359 |
For all the 38 INDELs, the genotype distributions were in agreement with Hardy–Weinberg equilibrium (HWE) expectations in the Jalisco state population, excepting G08 (p = 0.0475) (Table 1). However, after applying Bonferroni's correction for multiple tests (0.05/38 = 0.0013), all INDEL markers agree with the HWE expectations. Similarly, LD exact tests discarded allelic association between all pairwise combinations for the 38 INDEL loci (data not shown). In brief, these results (HWE and LD tests) validate the confident use of the binomial expansion of (p + q)2 and the product rule, respectively, to calculate the random matching probability (RMP) of DNA profiles based on HID-INDEL in forensic casework and kinship analyses.
ConclusionsBased on the analysis of 38 HID-INDELs in 25 Mexican families, we evaluated the proficiency provided by the genetic system to verify paternity through the PI. Important limitations arose when one parent was omitted from the kinship test, emphasizing the complementary value of this HID system. The comparison of PI, a posteriori kinship parameter, with the corresponding a priori parameter TPI, suggest that this should be redesigned for biallelic loci. Finally, we updated forensic parameters for the populations where the families come from (Jalisco state, Mexico).
The authors thank to all volunteers included in this study, and to the QFBs Alejandra Sandoval and Mariana Chávez for technical support. We also thank to the Mexican government by the financial support (grant CONACyT N° 286623) to H-R-V.