A simple and reliable method for diagnosing COVID 19 infections is the needed. The role of saliva in the transmission of the infection has already been established.
MethodSaliva and nasopharyngeal swabs from patients suspected to have COVID 19 infections were taken simultaneously, and the results of the RT-PCR were compared.
ResultTotal 405 samples were collected, of which 250 males and 155 females. In the 391 samples included for analysis, 370 (94.63%) samples were found to have concordance results, and 21 (5.37%) samples had discordant results.
ConclusionThe use of saliva to diagnose COVID 19 infection is reliable, and its use can be recommended.
Un método simple y confiable para diagnosticar infecciones por COVID 19 es necesario. Ya se ha establecido el papel de la saliva en la transmisión de la infección.
MétodoSe tomaron simultáneamente hisopos de saliva y nasofaríngeos de pacientes con sospecha de infección por COVID 19 y se compararon los resultados de la RT-PCR.
ResultadoSe recogieron 405 muestras, de las cuales 250 hombres y 155 mujeres. En las 391 muestras incluidas para el análisis, se encontró que 370 (94,63%) muestras tenían resultados de concordancia y 21 (5,37%) muestras tenían resultados discordantes.
ConclusiónEl uso de la saliva para diagnosticar la infección por COVID 19 es confiable y se puede recomendar su uso.
Coronavirus 2019 (COVID-19) infection due to the SARS-CoV-2 virus has proven to be a serious and deadly disease causing a great deal of mortality and morbidity globally.1 A key to preventing the spread of the virus is early diagnosis and isolation of infected individuals. Nasopharyngeal swabbing, followed by reverse transcription of the extracted RNA (RT-PCR), is the gold standard for detecting SARS-CoV-2 infection.2 This swab collection requires trained medical personnel3 and may not always be successful in the first attempt. In addition to exposing the healthcare personnel to a high risk of infection,4 they increase the consumption of personal protective equipment.2 The nasopharyngeal swab (NPS) collection method also causes discomfort to the patient5 and is difficult to perform in patients with nasal pathology like gross deviated nasal septum.6 Therefore there is a need for a simpler and less invasive method that reduces the risk to healthcare personnel and the economic and logistic burden on healthcare systems.
One potential candidate in this regard is saliva. Saliva collected from the mouth is a mixture of glandular secretions, gingival crevicular fluid, expectorated airway surface liquid and mucus, epithelial and immune cells from the oral mucosa and upper airways, and oral microbes and viruses.7 Saliva has proven useful in many oral and systemic conditions, including viral infections like dengue, West Nile, chikungunya, Ebola, Zika, and Yellow Fever.8–10 In addition, saliva has also been used for diagnosing coronaviruses responsible for severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS).11
Potential advantages of saliva specimens are that they can be easily be self-collected by adults and even children eliminating the need for health care professionals and reducing nosocomial infection. This will reduce the time and cost associated with the specimen collection. Easier sample collection will also help in increasing the compliance of patients and in mass screening. Considering these advantages, we planned a study to compare the efficacy of standard nasopharyngeal swabs and saliva in detecting the SARS CoV-2 infection.
MethodSample Size: Previous studies showed the saliva sample has a sensitivity of 85.19% compared with the oro-nasopharyngeal swab sample. Using this sensitivity and 95% confidential interval and 5% Precision, the sample size was calculated to be 194 using n-master 2.0 software.
The study was carried out in a tertiary care center in India from April to June 2021. The study was approved by the institute ethics committee (AIIMS/MG/IEC/2020-21/). After obtaining informed written consent, patients with symptoms of SARS-CoV-2 infection attending the sampling area of the hospital were taken for the study. Very sick or unstable patients with all forms of ARDS, sepsis, terminally ill, and patients using mouthwash regularly were excluded.
Saliva was collected by asking the patient to cough saliva from their throat into a sterile container, and 2 mL of viral transport medium was then added. They were asked not to eat, drink, smoke or use oral hygiene products for at least 10 min before the collection process. The nasopharyngeal swab was taken with flocculated swabs inserted into the nostril parallel to the palate just above the floor of the nasal cavity. The swab was inserted till resistance was felt and was left for a few seconds inside the nasal cavity. It was then gently removed by rotating it.
Saliva and Nasopharyngeal swab were collected from patients to detect SARS-CoV-2 by real-time reverse transcriptase RT-PCR. RNA was extracted from clinical samples with a QIAamp viral RNA mini kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. A final volume of 50 µl of RNA was eluted and kept at −80 °C till further processing. All specimens were handled with biosafety guidelines per the Indian Council of Medical Research (ICMR)/WHO/Center for Disease Control and Prevention for COVID-19.
After RNA extraction, one-step RT-qPCR (the reverse transcriptase reaction automatically precedes the PCR phase in the same tube) was performed using ICMR approved RT-PCR kitin Quantstudio 5 real-time PCR instrument (Thermo Fisher Scientific Inc.) based on the instruction for use (IFU) provided. Briefly, the master mix (a pre-mixed concentrated solution consisting of buffer, Reverse transcriptase enzyme, nucleotide, forward primer, reverse primer, TaqMan probe, DNA polymerase) was prepared dispensed into PCR tubes, and finally, RNA elutes were added. To check the validity of the test, negative control, positive control, and no template control were also added in each run. The basic cycling conditions were as follows: reverse transcription at 55 °C for 5 min, denaturation at 95 °C for 5 min, and then 45 cycles of amplification at 95 °C for 5 s and at60 °C for 15 s. Results were seen on the Quant studio 5 software. Each reaction was read for the screening and confirmatory genes after confirming the performance of positive control and negative control results. As suggested in the manufacturer's instruction, a proper sigmoid curve with a Ct value is taken as positive.
Statistical analysisThe agreement between both the tests was summarized as percent agreement and positive percent agreement; Kappa statistics were used to measure the association between types of sample collection. The Ct value of RT-PCR was compared using spearman correlation. A P-value of <0.05 was considered statistically significant. Analysis was done using R software.
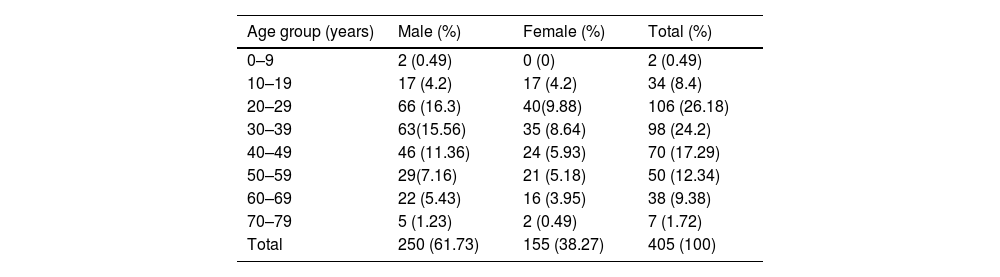
ResultA total of 405 samples were collected, of which 250 males and 155 females. The detailed age group distribution is presented in Table 1. Both nasopharyngeal swab (NPS) and saliva were processed. The sample is reported positive if either showed a positive sigmoid curve in the real-time reverse transcriptase PCR. Out of 405 samples, 159 NPS and 148 salivary samples were positive. The analysis did not include six NPS and eight salivary samples as they failed to show any fluorescence in the internal control.
Age wise distribution of the samples (n = 405).
| Age group (years) | Male (%) | Female (%) | Total (%) |
|---|---|---|---|
| 0–9 | 2 (0.49) | 0 (0) | 2 (0.49) |
| 10–19 | 17 (4.2) | 17 (4.2) | 34 (8.4) |
| 20–29 | 66 (16.3) | 40(9.88) | 106 (26.18) |
| 30–39 | 63(15.56) | 35 (8.64) | 98 (24.2) |
| 40–49 | 46 (11.36) | 24 (5.93) | 70 (17.29) |
| 50–59 | 29(7.16) | 21 (5.18) | 50 (12.34) |
| 60–69 | 22 (5.43) | 16 (3.95) | 38 (9.38) |
| 70–79 | 5 (1.23) | 2 (0.49) | 7 (1.72) |
| Total | 250 (61.73) | 155 (38.27) | 405 (100) |
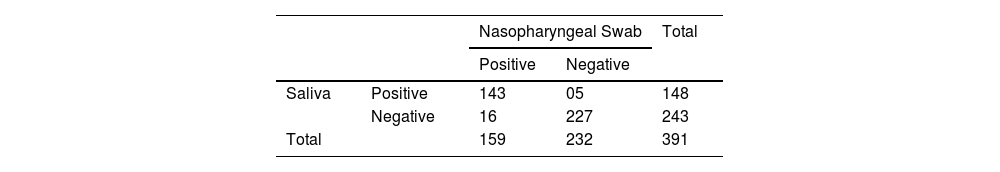
Out of 391 samples included in the analysis, 370 (94.63%) samples were found to have concordant results, and 21 (5.37%) samples had discordant results. Taking the nasopharyngeal swab as the gold standard sample, the saliva was compared with a 2 × 2 chi-square table (Table 2) with the online software epitools (https://epitools.ausvet.com.au/). The sensitivity of the saliva sample was found to be 89.94% with a 95% confidence interval (84.17–94.14) and specificity 97.84% with a 95% confidence interval (95.04–99.3) with a p-value of 0.02.
Taking a prevalence of 20% in the state of Andhra Pradesh,12 the positive and negative predictive values were calculated and found to be 91.25% and 97.49%, respectively, with an accuracy of 96.26%. The likelihood ratio (LR) was calculated and found to be 41.73 and 0.10, respectively, for positive likelihood ratio (LR+) and negative likelihood ratio (LR −).
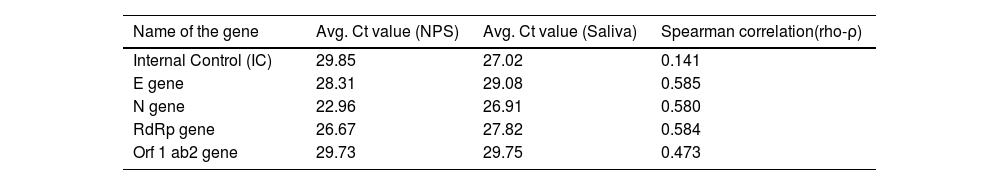
The threshold cycle value (Ct value) for both the samples in case of a positive result was also compared for various target genes like internal control, E gene, N gene, RdRp gene, and orf one ab2 genes (Table 3). All the parameters showed positive correlation with the rho (ρ) value ranging from 0.141 to 0.585.
Ct value comparisons of various genes in nasopharyngeal swab & saliva.
| Name of the gene | Avg. Ct value (NPS) | Avg. Ct value (Saliva) | Spearman correlation(rho-ρ) |
|---|---|---|---|
| Internal Control (IC) | 29.85 | 27.02 | 0.141 |
| E gene | 28.31 | 29.08 | 0.585 |
| N gene | 22.96 | 26.91 | 0.580 |
| RdRp gene | 26.67 | 27.82 | 0.584 |
| Orf 1 ab2 gene | 29.73 | 29.75 | 0.473 |
The use of saliva in diagnosing COVID 19 infections presents an exciting opportunity since it is easy to collect, non-invasive, and reduces the exposure and risk for healthcare workers involved in collecting the specimens. The use of saliva as a diagnostic modality has been reported in previous studies, with most of the studies supporting its role13–15 and some casting doubt on its usefulness.16,17
In our study, the sensitivity of saliva was 89.94%, which is higher than the previously reported sensitivity of saliva samples ranging from 83% to 86%18,19; however, these studies had a limited number of samples. A sensitivity of 100% has also been reported in a study20; however, only hospitalized COVID 19 were enrolled in this study. Rao M et al.13 found that the sensitivity and specificity of random saliva were higher than Nasopharyngeal + Oropharyngeal swabs (95.0; 99.9 vs. 72.2; 99.4). The variation in disease severity, the procedure of saliva collection, and the use of different target genes for detection may cause the variation in sensitivities reported in these studies.
In their study, Johnson AJ21 et al. found that saliva retained high clinical sensitivity even in the early pre-symptomatic stage of the disease. Other studies have also highlighted the efficacy of saliva in detecting the virus in the early stages of the disease.22,23 In addition, previous studies that evaluated the performance of saliva compared to nasopharyngeal swabs in children have also shown favorable results.24,25
The method of collection of saliva has also been reported in previous studies,26,27 and various techniques like early morning self-collection, coughing or deep throat saliva, and avoiding food, drink, or tooth brushing resulted in >5% increased positive saliva detection rates.28 Our study also obtained self-collected deep throat samples.
The majority of the studies have been performed in patients with severe COVID 19 infections or hospitalized patients; the advantage of our study is that the population studied was primarily mildly symptomatic or asymptomatic (contacts of confirmed COVID 19 cases). This shows the feasibility of using saliva on a larger scale for mass screening in the community. The self-collected non-invasive sample collection technique may make the population more permissible for testing and enhance community surveillance. Another advantage of our study is that we collected both the nasopharyngeal and saliva samples simultaneously from each participant; this will reduce the chance of variation in viral load among the specimens.
There are certain limitations of our study. Firstly our sample size is limited, and the setting is hospital-based. Further studies on community collected samples can be planned to assess the actual feasibility of collecting, transporting, and processing saliva samples.
ConclusionSaliva can be considered a reliable and efficacious alternative for nasopharyngeal swab specimens to detect COVID 19 infection. It is postulated that the use of saliva will reduce the risk of transmission of infection to healthcare workers, which will reduce the healthcare worker burnout seen during the pandemic. In addition, the demand for personal protective equipment and swabs will also reduce, thereby decreasing the cost of testing. The feasibility of using saliva in community settings should be explored further. In addition various aspects like asymptomatic presentation, severity of disease, positive history of contact, paediatric vs. adult, late vs. early course of disease should be evaluated in further studies as they may influence the results.
Conflict of interestNone.