The production and consumption of oysters is increasing annually because it can provide essential nutrients and benefit for human health, leading to frequent occurrence of severe allergic reactions observed in sensitized individuals. The aim of the present study was to investigate the effects of acid and protease treatment on the conformation and IgE-binding capacity of recombinant Crassostrea gigas tropomyosin (Cra g 1).
ResultsUnder acidic conditions, Cra g 1 did not undergo degradation, however, the changes obvious in the intensity of CD signal and ANS-binding fluorescence were observed, which was associated with a decrease in antibody reactivity. In simulated gastrointestinal fluid (SGF) and simulated intestinal fluid (SIF) digestion system, acid-treated Cra g 1 was relatively resistant to digestion, but the degradative patterns were very different. Moreover, owing to alterations of secondary structure and hydrophobic surface of the protein during digestive processing, antigenicity of acid-induced Cra g 1 reduced in SGF while it increased significantly in SIF.
ConclusionTo our knowledge, this is the first study reporting that antigenicity of acid-treated oyster tropomyosin increased after SIF digestion. These results revealed that treatment with acid and pepsin, rather than trypsin, was an effective way of reducing IgE-binding capacity of tropomyosin from oyster.
Allergic diseases have long been considered an important health issue in the world. Food allergy is a typical disease that belongs to a type-I allergic reaction mediated by IgE.1 It is estimated that about 3–8% of children and up to 3% of adults are affected by food allergies.2 Among all food allergies, shellfish allergy is one of the most common types with a prevalence of 0.3% in the world population. Oyster, one of the typical representatives of shellfish, provides essential nutrients and benefits for human health. With the increase in production and consumption of oyster, the allergic diseases caused by it have also increased. Sensitized individuals exposed to oyster may suffer a series of immediate allergic reactions, which are characterized by gastrointestinal diseases, vascular plaques, itchy throat and even life-threatening anaphylaxis.3,4 To date, six shrimp allergens have been identified, including tropomyosin, myosin light chain, arginine kinase, troponin C, sarcoplasmic calcium binding protein and triose phosphate isomerase,5–7 whereas tropomyosin is the only well-identified allergen in oyster.4 Ishikawa et al.8 isolated the oyster major allergen, tropomyosin, from Crassostrea gigas for the first time and named it Cra g 1. Experiments have demonstrated that tropomyosin, a coil-coiled dimeric protein with a molecular weight of about 34–38kDa, is a highly conserved actin-binding protein in muscle.9–13
Some high-acid commercial products containing oyster residues, such as salad dressings, are increasingly consumed. In these products, organic acids, like vinegar, may affect the physicochemical properties of protein.14 The treatment of acids may produce some chemical modifications to allergenic proteins, which would eventually lead to changes in the conformation and allergenicity of them.15–17 In addition, simulated gastrointestinal fluid (SGF) and simulated intestinal fluid (SIF) digestion assay system are considered to be an important method to estimate food allergy.18 Astwood et al.19 performed simulated digestion experiments on common allergens and non-allergenic proteins, and found that allergenic proteins were highly resistant to digestion while non-allergenic proteins were quickly degraded. But some opposite results have been achieved in other literature, showing that allergenic and non-allergenic proteins have similar resistance to digestion.20,21 So far, there have been few reports on the stability, conformation and IgE-binding capacity of acid-treated oyster tropomyosin and its digests by protease. Hence, studies on the alterations of structure and immunoreactivity of tropomyosin during such processing are needed to ascertain the effects of acid and protease on its allergenic potential.
Therefore, the aim of our present study was to express recombinant Cra g 1 in Escherichia coli strain BL21 (DE3) and investigate the effects of acid and protease treatment on the conformation and IgE-binding capacity of this protein. These results will be useful to reduce the risk of foods containing oyster ingredients for oyster-allergic subjects.
Materials and methodsHuman seraSerum samples from seven oyster-allergic subjects were obtained from PlasmaLab International (Everett, WA, USA). All seven subjects had a clinical history of at least two or more symptoms within 1h of oyster intake, including pruritus, allergic rhinitis, gastrointestinal disease and diarrhea. A serum pool from these patients was made by mixing equal aliquots of IgE. Negative control sera were obtained from three subjects without adverse reactions after ingestion of shellfish. All sera were stored at −80°C until used. All experiments were approved by China National Research Institute of Food and Fermentation Industries (Approval number: 2017–0012).
Cloning, expression and purification of recombinant Cra g 1According to the Cra g 1 gene sequence (NCBI accession number AB444943.1), one pair of primers was designed: P1: 5′-CGGAATTCTGGACAGCATCAAGAAGAA-3′, P2: 5′-TATTGCGGCCGCTTCTTATTATTTTTCCATGGG-3′. The underlined portions are the restriction enzyme sites for EcoRⅠand NotⅠ. Then the Cra g 1 gene was amplified and inserted into the pET-21a vector. The resulting plasmid was verified by restriction digestion and sequencing from both ends of the inserted segment.
The recombinant plasmid was transformed into BL21 (DE3), plated on ampicillin-containing plates, and cultured overnight at 37°C. Single colonies were inoculated into 20mL of ampicillin-containing LB medium overnight at 37°C with shaking. 10mL of overnight culture was transferred to 1L fresh LB medium and grown at 37°C. When the OD600nm reached 0.6–0.8, 0.5mM IPTG was added to induce expression. After culturing at 18°C for 18h, the cells were harvested by centrifugation, resuspended in solution (20mmol L−1 Tris-HCl pH 8.0) and ultrasonicated (on for 3s, off for 3s, 99 cycles). Then, it was filtered through a 0.22μm filter after centrifugation at 15000rpm for 30min. The filtered protein solution was loaded onto a Ni2+ charged HiTrap chelating HP column of an ÄKTA-fast protein liquid chromatography (FPLC) system. After washing the column, the bound Cra g 1 was eluted with a linear 20–500mmolL−1 gradient of imidazole in the same buffer. To further purify the protein, the concentrated elution was applied onto a gel-filtration Hi-load Superdex-75 column equilibrated with a buffer containing 20mmolL−1 Tris-HCl, pH 7.5, 150mmolL−1 NaCl and 1mmol L−1 DTT. All purification procedures described here were performed at 4°C. The purified Cra g 1 was analyzed on 15% SDS-PAGE and stored at −80°C until used.
Protein quantificationThe total protein content of the purified extract was determined using the Quick Start Bradford Assay kit (Bio-Rad, Hercules, CA, USA) in accordance with the manufacturer’s instructions. Bovine serum albumin was used as the protein standard.
Acid treatment of Cra g 1The purified recombinant tropomyosin was treated under acidic condition to study the effects of different pH values and induction time on Cra g 1. The specific procedure was as follows: 0.5mgmL−1 of Cra g 1 was treated at three pH gradients at pH 5.0, 3.5 and 2.0 for 1 and 3h, respectively. Cra g 1 without acid treatment was used as control.
SGF digestion assaySGF was prepared as described in the United States Pharmacopoeia.22 SGF consists of porcine pepsin (275 U/mg) in 25mmolL−1 NaCl with pH 2.0. For digestion assays, a centrifuge tube (10mL) containing SGF was preheated at 37°C prior to addition of test protein to adequately simulate human gastric fluid temperature. A ratio was determined (1:100 pepsin to protein, w:w) for tests at five different time points (0, 5, 15, 30, and 60min). The digestion was performed at 37°C and terminated by adding 200mmolL−1 Na2CO3. The protein sample for 0-minute time point was added to SGF and immediately stopped with Na2CO3. For the control, the protein sample was mixed with SGF without pepsin. All the experiments were performed in triplicate.
SIF digestion assaySIF was prepared as described in the United States Pharmacopoeia and was composed of trypsin in 0.05molL−1 KH2PO4 with pH 7.5.22 For digestion assays, a centrifuge tube (10mL) containing SIF was preheated at 37°C prior to the addition of test protein. For final digestion, a ratio (1:100 trypsin to protein, w:w) was selected for all tests at six different time points (0, 15, 30, 60, 120, and 240min). The tests were carried out at 37°C and heated at 95°C for 5min to terminate reaction. At the 0-minute time point, the protein sample was added to SIF and immediately heated at 95°C for 5min. For the control, the protein sample was mixed with SIF without trypsin. All the experiments were performed in triplicate.
Electrophoresis analysis (SDS-PAGE)The sample solution (20μL) was mixed with 5μL of the loading buffer, heat-denatured, and loaded into each well of the SDS-PAGE gel (5% stacking gel, 15% running gel). The molecular weight was determined by comparing the electrophoretic mobility of molecular weight marker proteins (14.4–116kDa) purchased from Takara Biotechnology Company (Dalian, China).
Circular dichroism (CD) spectroscopyThe CD spectrum of the protein sample (0.3mgmL−1) was measured with a Jasco J-810 Spectropolarimeter (Jasco Inc., Easton, MD, USA) and the average of three spectra was taken as the final data. For wavelength analysis, samples were scanned with step increments of 0.2nm and a band width of 2.0nm. The spectral range was 190–250nm and the scanning speed was 200nmmin−1. The baseline spectrum of buffer was subtracted from each spectrum and the resulting value was converted to molar ellipticity (θ) using the analysis function built into the Jasco software.
ANS-binding fluorescence measurementsThe hydrophobic surface was monitored by ANS-conjugated fluorescence using an F-4500 fluorescence spectrophotometer (Hitachi Inc., Tokyo, Japan). 1.0mL of different samples were mixed with 15μL of 5.0mM ANS solution and reacted for 30min at 25°C. Both the excitation and emission slit widths were 5nm and the scan speed was 1200nmmin−1. 300μL aliquots of each sample were added to a quartz cuvette with 1cm path length. Experiments were performed in triplicate and their average values were taken as the final results.
IgE enzyme-linked immunosorbent assay (ELISA)The samples were diluted to 5μgmL−1 with CBS solution, 100μL of each sample was taken to a 96-well plate (Costar Inc., Cambridge, MA, USA) and coated at 4°C overnight. After washing four times with Tween 20/PBS (PBS-T), 350μL of 5% skimmed milk powder was added and incubated at 37°C for 1h. It was washed and 100μL of pool of sera from allergic patients diluted (1:50) in 2% skimmed milk powder was added and incubated for 3h at room temperature. After washing, 100μL of peroxidase-labeled goat anti-human IgE antibody (1:1000;KPL Inc., Gaithersburg, MD, USA) was added to wells and then plates were incubated at room temperature for 1h. After washing five times with PBS-T and three times with PBS, 200μL of TMB solution was added to wells for 30min, 50μL of 2M H2SO4 was used to stop the reaction. Finally, the O.D. values were measured with a spectrophotometer (Dynex TechnologiesInc., Chantilly, VA, USA) at 450nm. The same procedure was performed with sera from non-allergic subjects to determine the extent of non-specific binding, which was subtracted from test sera data. Three parallel experiments were performed for each sample and the average values were taken as the final data.
Statistical analysisData were processed using Origin 8.0 software (OriginLab Corp., Northampton, MA, USA). All quantitative data for the experiments were mean±standard deviation. Significant difference in means between the samples was determined at a 5% confidence level (P<0.05).
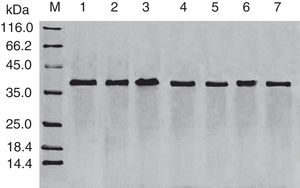
Results and discussionExpression and purification of Cra g 1The DNA fragment encoding Cra g 1 was cloned into expression vector pET-21a and confirmed by restriction enzyme digestion and DNA sequencing. The protein expression was carried out in E. coli BL21 (DE3). Cells were collected and analyzed by SDS-PAGE. After two-step purification by Ni2+ affinity chromatography and S-75 gel-filtration chromatography, the recombinant protein was purified to near homogenous (Fig. 1, lane 1). The final yield of this protein is 1mgmL−1 and the purified protein was stable after storage at −80°C.
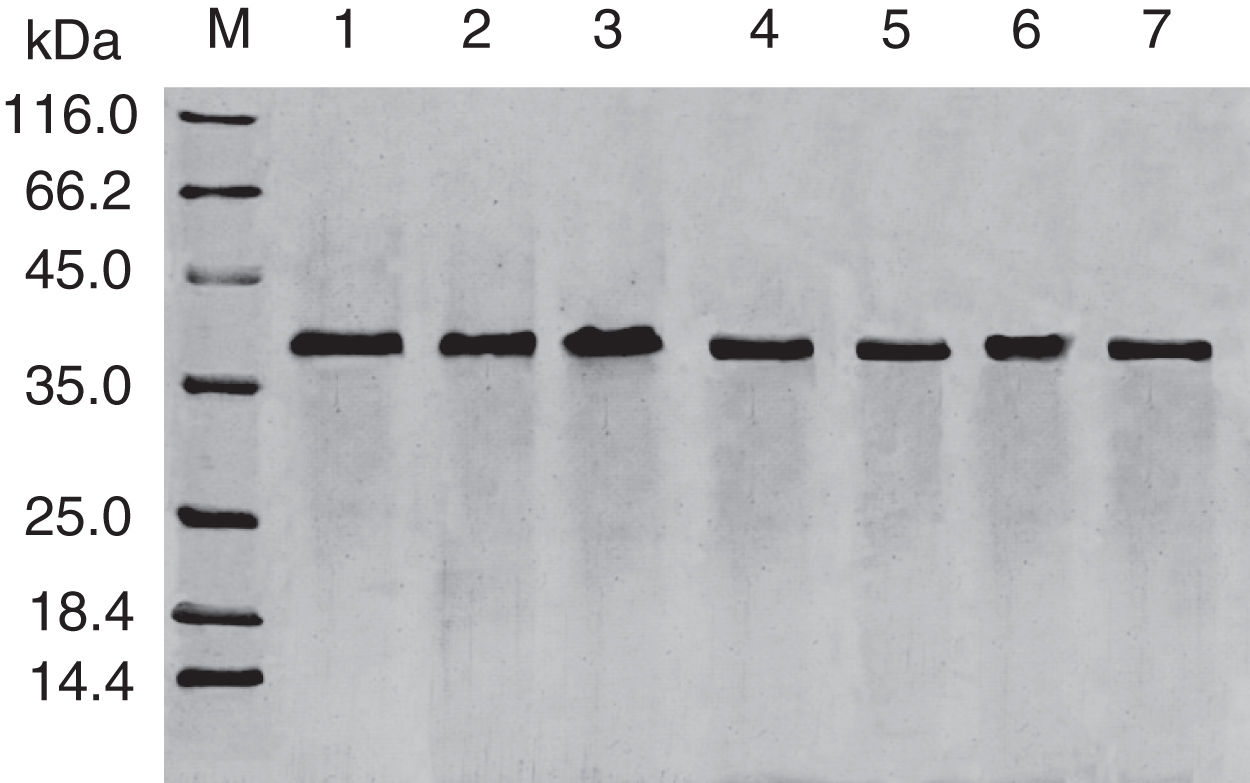
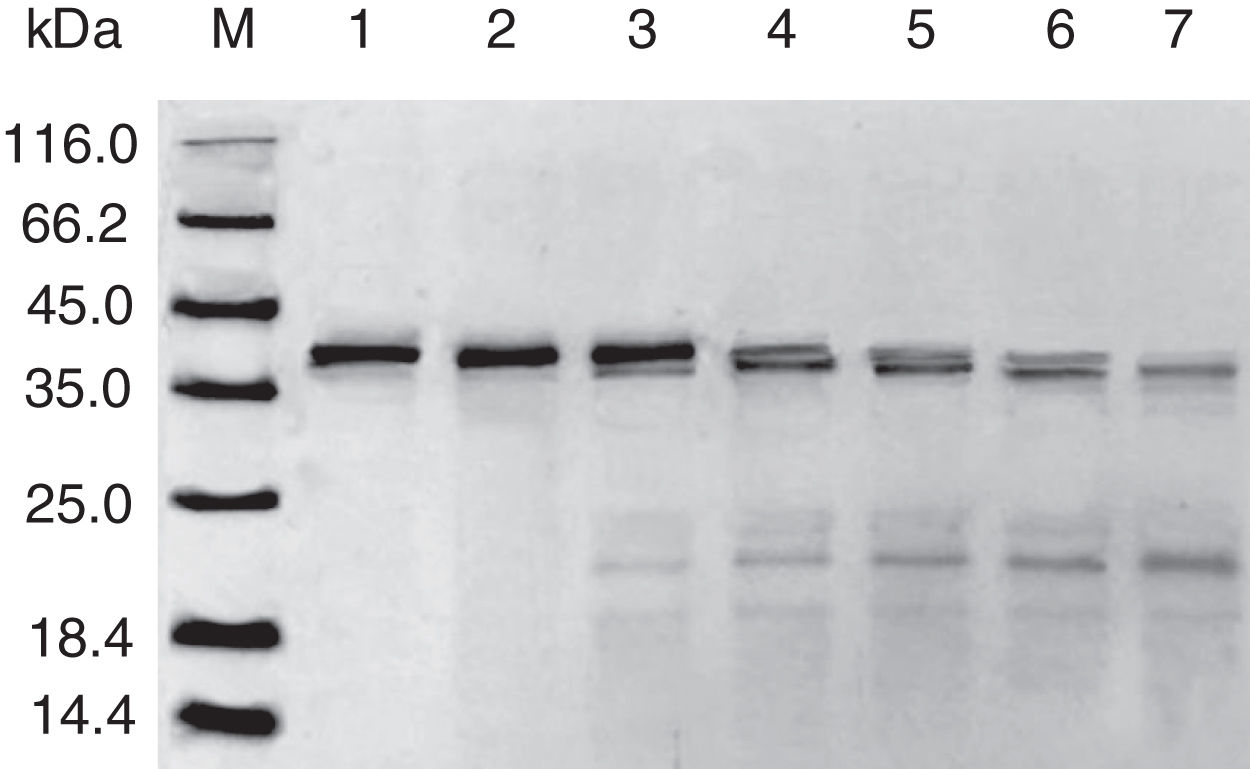
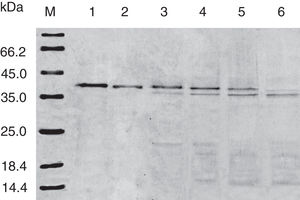
SDS-PAGE analysis of effect of acid on recombinant Cra g 1. Lane M, molecular mass marker (kDa);Lane 1, purified Cra g 1; Lanes 2–4, Cra g 1 was treated by acid for 1h at pH 5.0, 3.5 and 2.0, respectively; Lanes 5–7, Cra g 1 was treated by acid for 3h at pH 5.0, 3.5 and 2.0, respectively.
The SDS-PAGE of Cra g 1 treated with different pH and time was shown in Fig. 1. After acid treatment at pH 5.0, 3.5 and 2.0 for 1h and 3h, an apparent protein band of approximately 37kDa was still observed (Fig. 1, lanes 2–7), and did not change under acidic conditions. Almost no degradation produced low molecular weight protein bands.
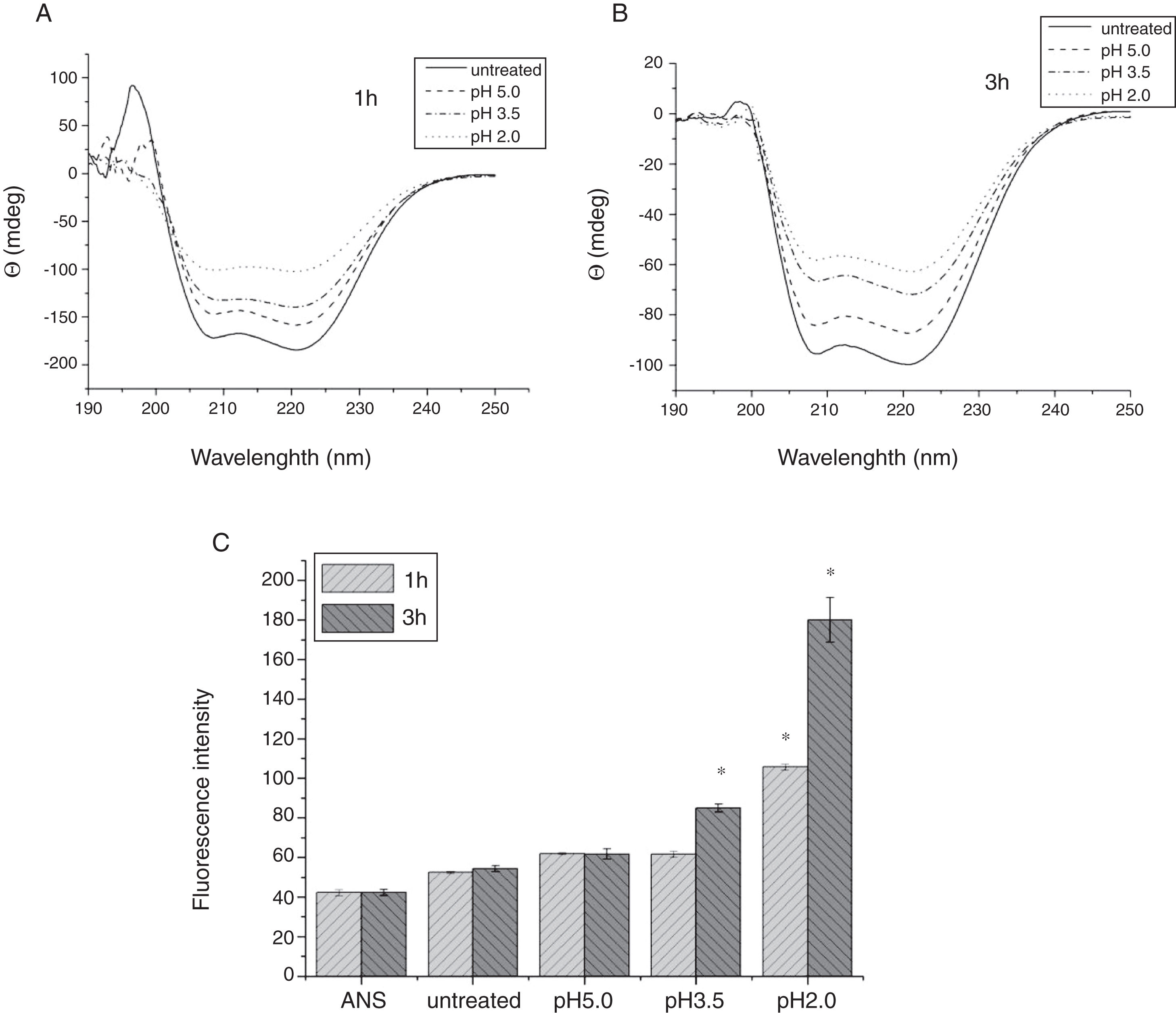
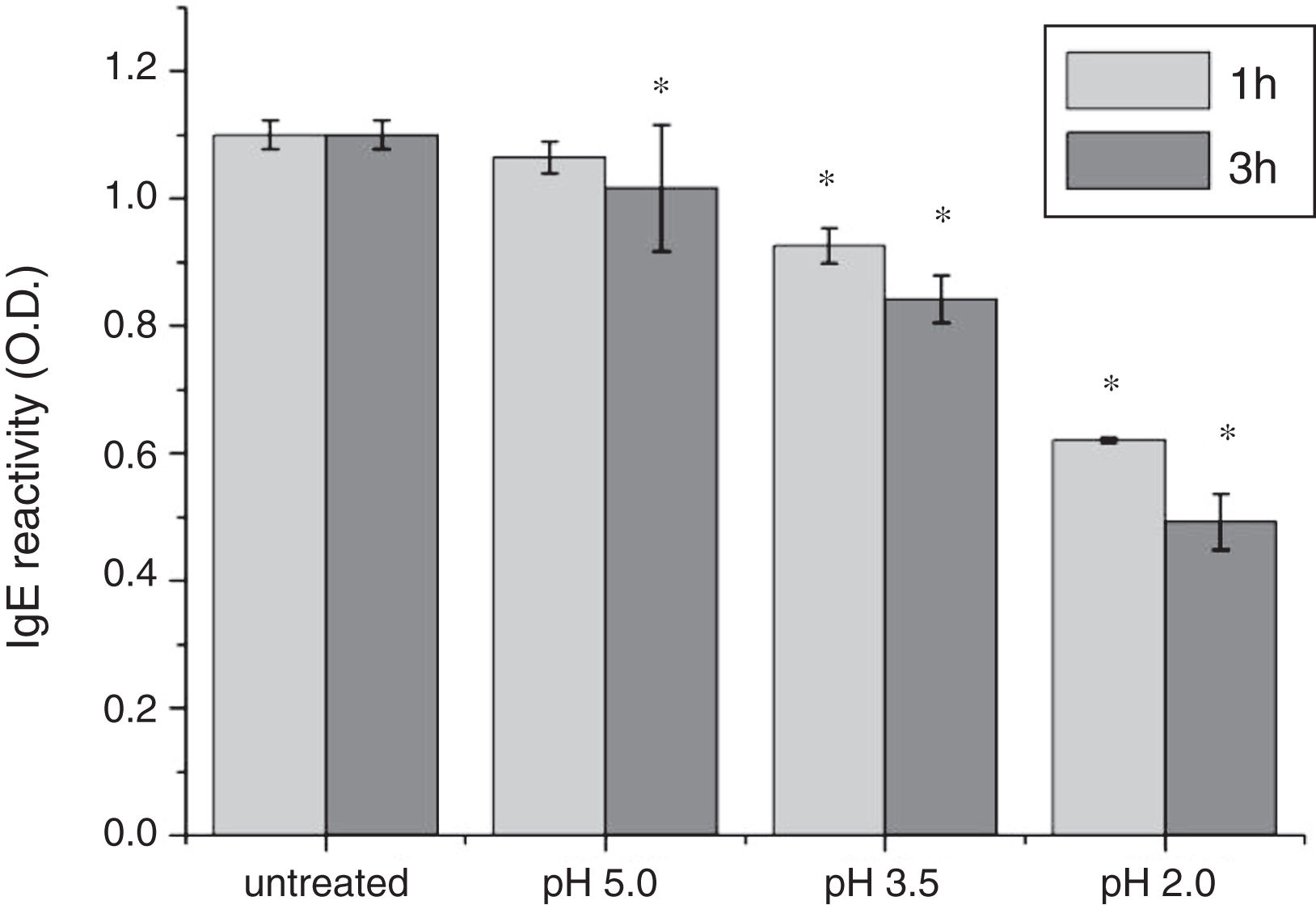
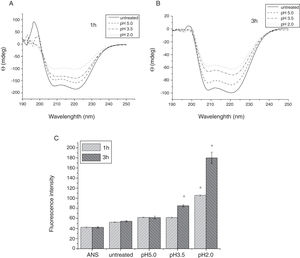
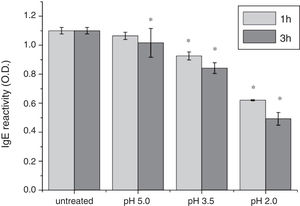
To analyze acid-induced changes in the structure of Cra g 1, CD spectra measurements were allowed to monitor the folding changes. The CD spectrum has a peptide bond absorbing signal in the far-UV region and reflects the contents of the regular secondary structure feature of protein. From our CD spectra (Fig. 2A and B), we found that the original Cra g 1has a positive peak (maximum) at nearly 198nm and two negative peaks (minimum) at nearly 208nm and 222nm. Moreover, the molar ellipticity 222/208 ratio is greater than 1. These results indicated that it is a typical α-helical protein and forms a double-stranded coil-coiled structure. On the other hand, significant changes in the intensity of signal could be observed after acid treatment at pH 5.0, 3.5 and 2.0 for 3h. When the pH ranged from 5.0 to 2.0, the increase in the intensity, coupled with the minimum, was shifted to a lower wavelength, indicating that α-helix content of tropomyosin was further reduced and coil-coiled structure was partially damaged. The results of acid induction for 1h were not as obvious as those of 3h. Next, ANS-binding fluorescence was used to determine the hydrophobicity of Cra g 1 after acid processing. The surface hydrophobicity of acid-treated Cra g 1 was increased with pH environments ranging from 5.0 to 2.0 for 1h or 3h. Surprisingly, after 3h of acid treatment at pH 2.0, the surface hydrophobicity of Cra g 1 was almost three times compared to that of the original form (Fig. 2C). The IgE-binding capacity of acid-induced tropomyosin was evaluated by indirect ELISA (Fig. 3). As we speculated, the untreated Cra g 1 showed higher IgE reactivity, however, acid-treated tropomyosin reduced IgE-binding capacity when exposed to low pH environments, particularly at pH 2.0 and 3.5 for 1h and 3h (P<0.05).
Conformational changes of Cra g 1 before and after acid treatment. (A) CD spectra of Cra g 1 treated with acid for 1h at different pH; (B) CD spectra of Cra g 1 treated with acid for 3h at different pH; (C) ANS-binding fluorescence of Cra g 1 treated at different pH and time. *P<0.05 vs. untreated protein. All data are presented as the mean±SD (n=3).
Our results illustrated that Cra g 1 did not undergo degradation under acidic conditions, indicating that it was resistant to acid, which agreed well with previous reports.17 In addition, the conformation of the protein altered greatly during processing, which suggested that the secondary structure was partially destructed and more hydrophobic regions were exposed on the surface of Cra g 1. Generally, ionic interactions and hydrophobic interactions are considered to be the most important factors in forming the three-dimensional structure of protein.23 Acids could affect the protonation state of the charged amino acid, alpha-carboxyl, alpha-amino terminal groups as well as hydrophobic surface of protein, which may ultimately alter the possible IgE-binding epitopes and potential allergenicity. Thus, we might deduce that steric structure of Cra g 1 was changed under acidic conditions, leading to destroying or modifying several antigenic epitopes on molecular surface. These changes markedly reduced IgE reactivity compared to the original protein, which is consistent with previous experimental results.24 In order to further explore the conformational and allergic alterations of acid-treated Cra g 1 during human digestion, we finally selected the protein that was to deal with acid for 3h at pH 2.0 to use in simulated gastrointestinal fluid digestion experiments and the digestion time in SGF and SIF was designated as 1h and 4h, respectively, based on the detention time of foods in digestive system.
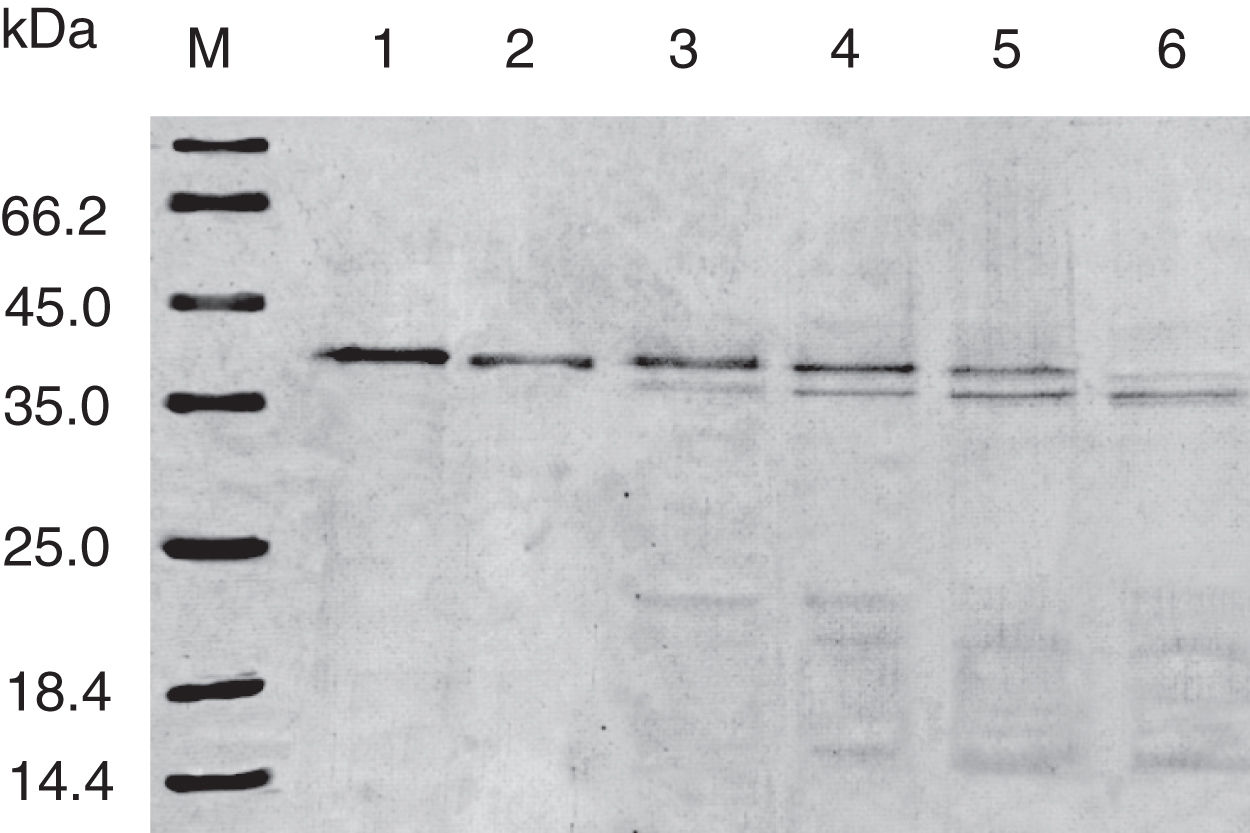
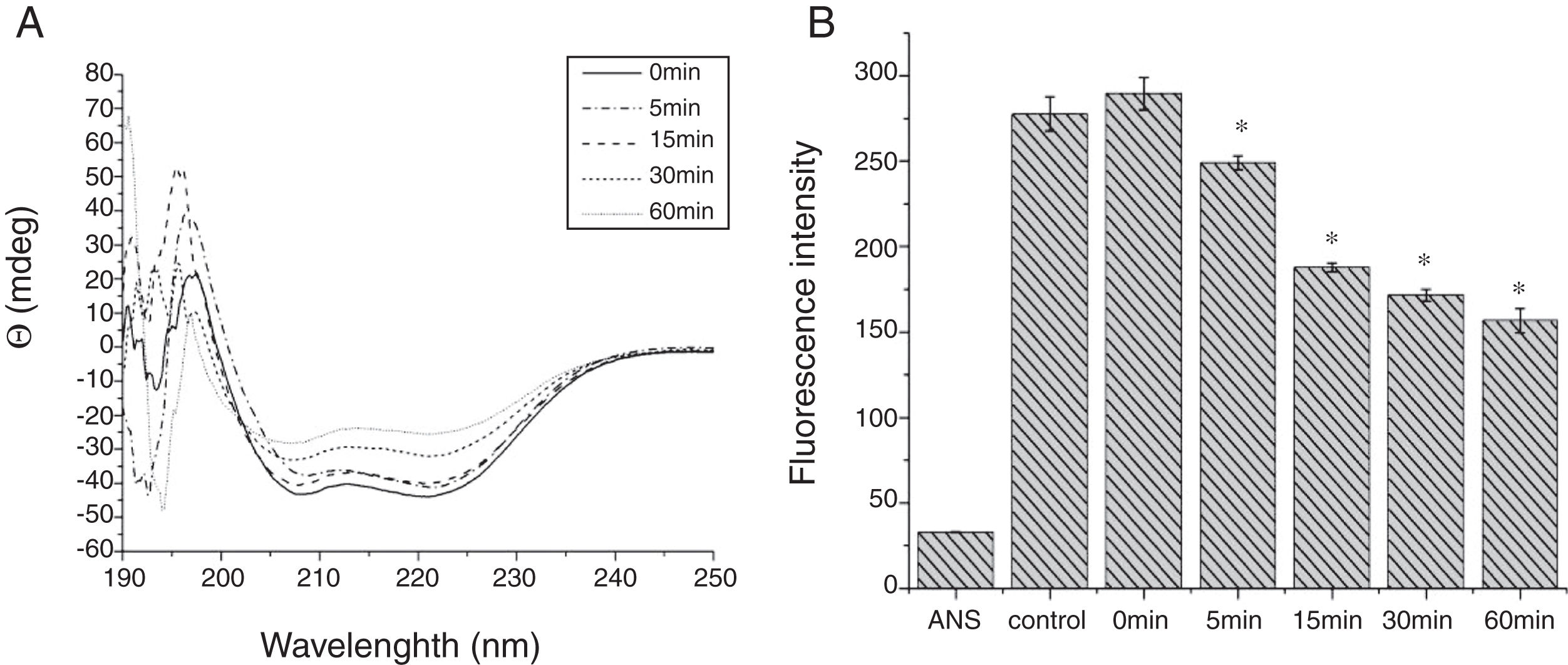
Digestibility of acid-treated Cra g 1 by SGFIn the present study, acid-treated Cra g 1 was subjected to SDS-PAGE analysis after SGF digestion (Fig. 4). Cra g 1 was relatively resistant in SGF and was only slightly degraded even at higher digestion periods. Many digestion-resistant fragments (about 36, 22 and 15kDa) progressively intensified with prolongation of digestion time, while intact protein did not completely degrade at 60min by pepsin.
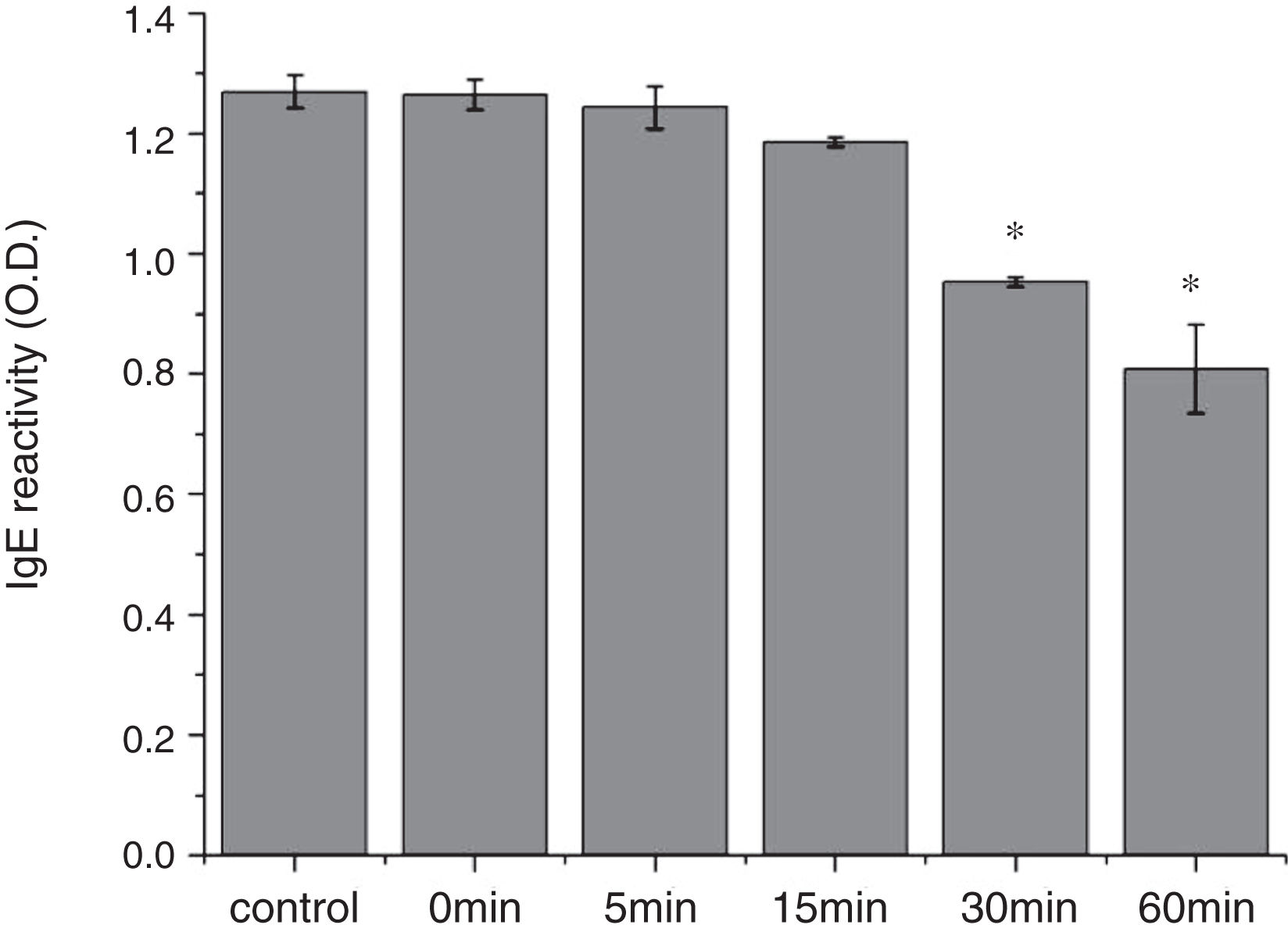
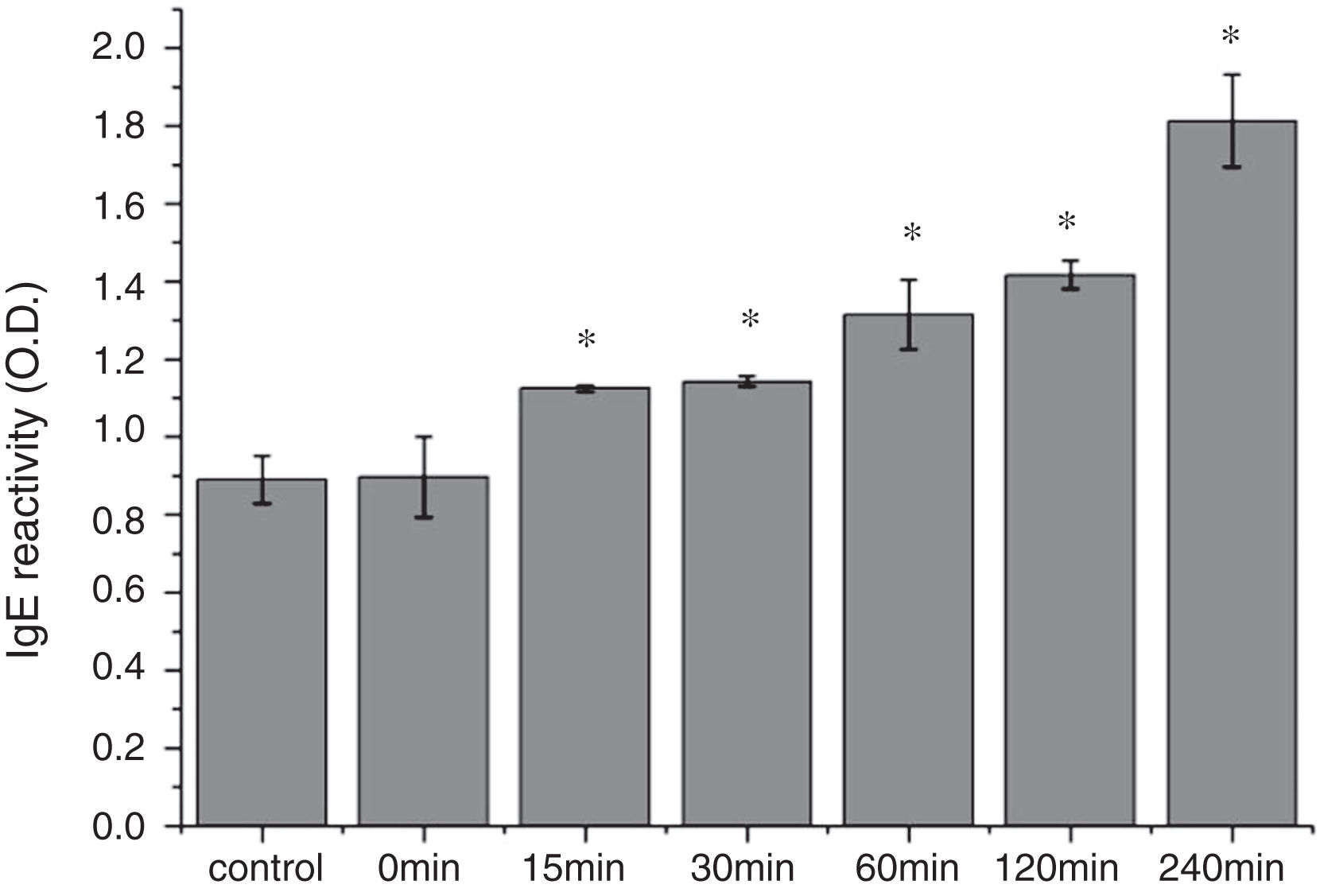
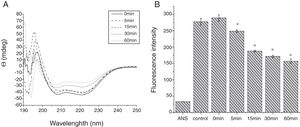
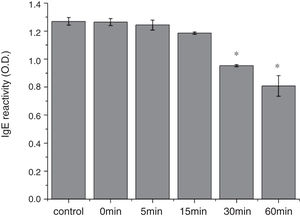
From the CD spectra, the signal obviously changed after SGF digestion (Fig. 5A). With increasing digestion time, the intensity of CD signals was increased and the minimum gradually shifted to lower wavelengths, implying the secondary structures were varied drastically. Hydrophobicity of Cra g 1 after pepsin treatment was determined by ANS-binding fluorescence measurements. Interestingly, compared to the control, the surface hydrophobicity of the protein was decreased by 4%, 27%, 34%, and 40% when digestion time was adjusted from 5 to 60min (P<0.05) (Fig. 5B). To further analyze whether SGF digestion affects the allergenicity of acid-treated Cra g 1, we performed an indirect ELISA experiment using pooled oyster-allergic subjects’ sera. Compared with the undigested sample, IgE-binding ability showed a decreasing trend from 5 to 60min, especially at 30 and 60min (P<0.05) (Fig. 6).
Pepsin is the only proteolytic enzyme in the stomach, which prefers to cleave peptide bonds following Phe or Tyr residues, as well as other hydrophobic amino acids.25 Pepsin could only slightly hydrolyze oyster tropomyosin, demonstrating that it has relative resistance to pepsin digestion (Fig. 4). A possible reason is that the structural changes of Cra g 1 caused by acid produced some modifications to the enzyme cleavage sites and fewer Phe and Tyr residues were exposed on the surface of the protein, thus diminishing the degree of hydrolysis. Additionally, we might infer that pepsin, at least in part, altered the conformation of the protein during digestive processing, resulting in hydrophobic residues being masked and several antigenic epitopes that bound serum IgE were buried inside the protein (Figs. 5 and 6). This phenomenon was also observed by Huang YY et al.;26 they demonstrated that mud crab (Scylla serrata) tropomyosin reduced IgE-binding ability after pepsin digestion.
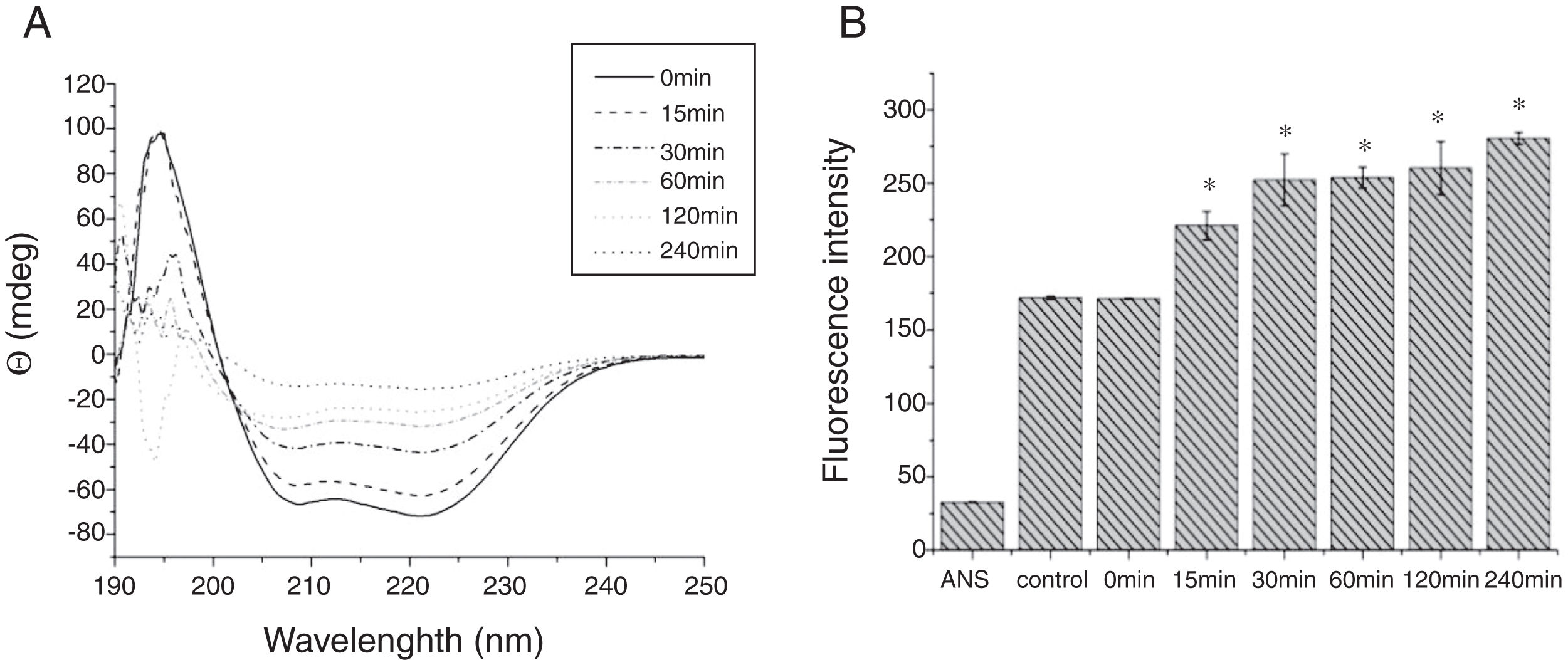
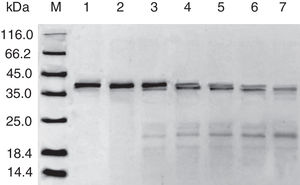
Digestibility of acid-treated Cra g 1 by SIFThe degradative patterns of acid-treated Cra g 1 by SGF and SIF were quite different. Under SIF conditions, more obvious degraded fragments appeared (Fig. 7). Cra g 1 was digested into three main proteolytic fragments with sizes of approximately 36, 22 and 20kDa. All these fragments and the intact original protein remained even after 4h by trypsin digestion.
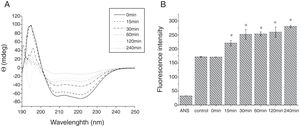
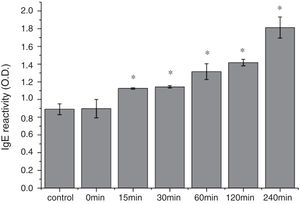
The CD far ultraviolet signal of acid-treated Cra g 1 after SIF digestion is shown in Fig. 8A. The spectra gradually showed a slight blue shift in the course of digestion, and the curve was nearly flat after 240min. These results demonstrated that the regular α-helices that dominate in tropomyosin structure diminished when the protein environment varied. Besides, compared with the undigested Cra g 1, the surface hydrophobicity increased by 28%, 43%, 46%, 53% and 58% after 15, 30, 60, 120 and 240min, respectively (P<0.05) (Fig. 8 B). The IgE-binding ability of Cra g 1 after SIF digestion was further analyzed by ELISA (Fig. 9). Surprisingly, contrary to the SGF results, all digested samples have significantly higher IgE reactivity than the control (P<0.05). Possibly, more IgE-binding epitopes in degraded fragments or original protein were generated by trypsin digestion. Therefore, this treatment may not be effective in decreasing the allergenic ability of the protein.
The sensitization of potential allergens is routinely evaluated using simulated digestion studies. Our results showed that digestive patterns by trypsin and pepsin were very different because of cleavage specificities of the two proteases. Trypsin mainly cleaves peptide bonds following Lys and Arg residues at the P1 position with higher specificity.27 Although there are several cleavage sites for trypsin and pepsin in Cra g 1 primary structure,28 acid-treated protein and its degraded fragments were resistant to digestion (Figs. 4 and 7). One reason for this might be that cleavage sites on the molecular surface were eliminated due to acid treatment. Hence oyster tropomyosin, as well as its digestion-resistant fragments, could survive and reach the intestinal immune system. Furthermore, antigenicity of acid-induced Cra g 1 decreased in SGF, while it increased significantly in SIF (Figs. 6 and 9). This phenomenon may result from conformational alterations of protein (Fig. 8), which exposed more hydrophobic groups and additional immunogenic epitopes by trypsin digestion. It is also possible that new IgE-binding epitopes were produced in proteolytic fragments by SIF, which contributed to the IgE-binding reactivity. To our knowledge, this is the first report that antigenicity of acid-treated oyster tropomyosin increased under SIF conditions. This explains why sensitized individuals still suffer severe allergic reactions even if they intake high-acid oyster products such as salad dressings. Besides, the ELISA data are a relative result because IgE-binding capacity does not equate to allergenicity completely.29 Some degraded fragments may bind IgE but not enable basophils or mast cells to release mediators. Consequently, the next steps are needed to assess the allergic changes of tropomyosin through release of histamine from basophils and skin prick test following acid and protease treatment. Identification of immunogenic epitopes in intact Cra g 1 and its proteolytic fragments during the same processing should be carried out to further expound the molecular mechanism of interactions between the protein and oyster-specific IgE.
In conclusion, alterations in secondary structure and exposure or burial of hydrophobic amino acid residues were responsible for conformational changes, resulting in the disruption or formation of IgE-binding epitopes, and ultimately affected immunogenic activity. For oyster tropomyosin, treatment with acid and pepsin, rather than trypsin, is an effective way of reducing IgE-binding capacity.
Conflict of interestThe authors have no conflict of interest to declare.
This work was supported by grants from the General Program of National Natural Science Foundation of China (No. 31671963), the National Key Research and Development Program of China (No. 2016YFD0400604), and the Beijing Municipal Science and Technology Program (No. Z161100005016030 and No. Z171100001317006).