INTRODUCTION
PubMed is one of the better tools to seek in the biomedical literature, with the only exception of certain very specific documents. One of its main advantages is the immediacy of the information offered, because is updated daily from Tuesday to Saturday. Furthermore it is an International Data base, although the nationality of the indexed papers is recorded this is not a criterion of selection.
It is estimated that 40 % of world's biomedical literature is included in PubMed, nevertheless is necessary to have a minimum knowledge on it, its operation and its limitations.
Pubmed can be used by anybody who has a computer connected to the web, without any limitation or registration requirement (the only exception being some special service) and without any cost, the only cost is that associated with the Internet Provider.
It is produced by the National Library of Medicine, NLM and the National Center for Biotechnology Information, NCBI, of the United States of America, both centers are governmental centers.
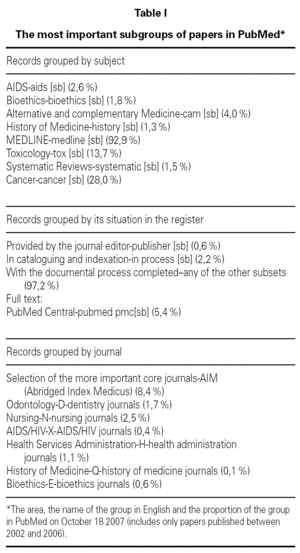
PubMed keeps records of biomedical papers published in scientific journals from many countries. These records are organized in subgroups or subsets see table I each of one can be consulted independently, it is possible also to consult simultaneously two o more subsets. The criteria for pooling the records in these subsets are the thematic contain or the methodological aspects of the papers, journal specialty, and its situation in the registration process that determine the amount of information provide. None of these subgroups is specifically dedicated to allergology or inmunopathology. Among these subgroups that are the final destination of the record lies Medline that contents more than 90 % of all the PubMed's records. Some records allow the access to the complete full text of the paper; those form the "PubMed Central" subset. There is an overlapping between groups, i.e. a record may be included in several subsets.
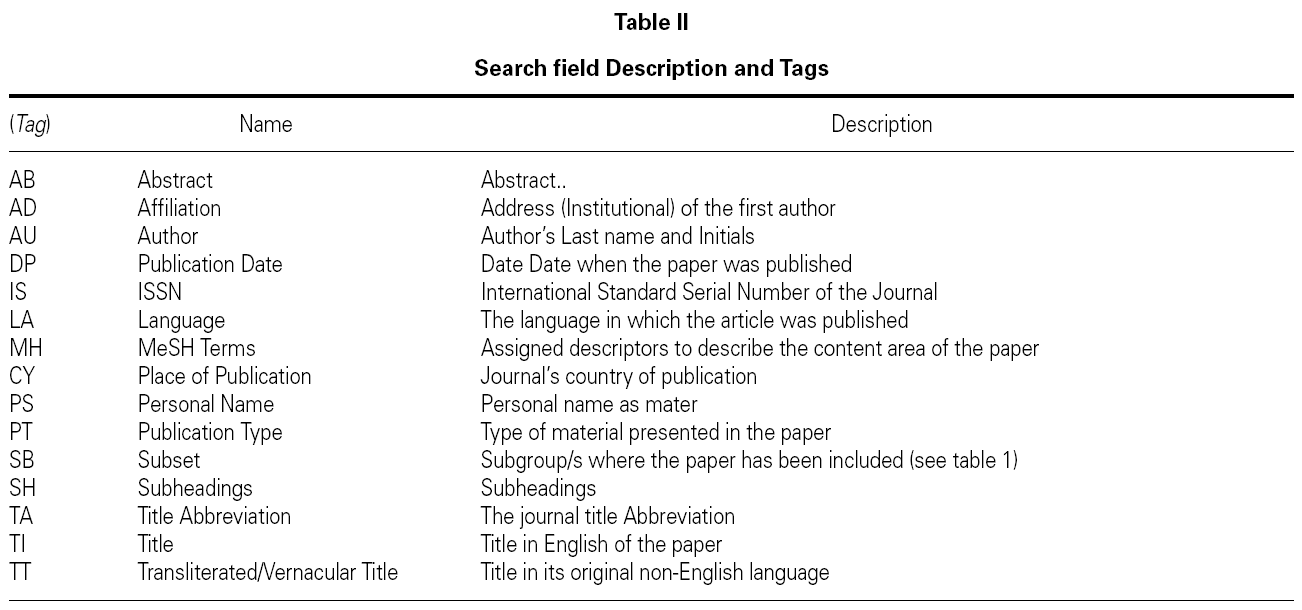
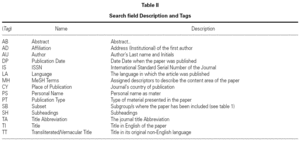
All the information in each of the records is organized in 'fields', which are identified bay a tag, usually the first better of the English name of the filed. Some of the more useful fields for searching information are presented with their tags in table II.
Now PubMed keeps more than 17 million records, they are progressively increasing, in the nineties the increase was 500000 new records per year, now the number is close to 700000 new records per year. Nevertheless whoever wants to use either PubMed or Medline must take into account the existence of limitations due to the selection process of the information included in the data base. In September 18th 2007 90.1 % of all the papers between 2002 and 2007 and registered in PubMed were written in English no other language represents more than 1.5 %.
The analysis by country of papers mentioned above shows that almost half of them appeared in journals printed in the US. Papers published in the 27 countries of the EU plus three candidate countries (i.e. 30 European countries), accounts for approximately 40 % of the records. Furthermore there are inequities among the EU countries because approximately half of the papers are from the UK.
GENERAL PRINCIPLES OF BIBLIOGRAPHIC SEARCH IN PUBMED
Let us begin thinking that the operation of bibliographic data bases, such us PubMed, is similar to Internet search engines (although there are some differences that will be presented later) in any case the process for retrieving information is always the same.
a)To determine what information is needed and what will be its use.
b)Based on it, we must establish what words either unique (e.g. Hypertension) or composed (as diabetes mellitus) are related to that information.
c)Enter the significant terms into the search box to ask the search engine or data base to look for the information.
This work system consists in a search of information in natural language. That means that we are using to communicate with the software the same language which we use to communicate with other people, this has many advantages but also some disadvantages. Among these we must emphasize that the natural language has a high variability (e.g. gender and number), the existence of synonymy (two words or phrases are synonyms when they have the same meaning) and polysemes (a word or a phrase that can be used to express two or more different manings.
PubMed is not an exception and can be consulted using natural language. All we need is to determine the correct form of the information that we want, what words are related with that and to write them in the search box. From now, these words from the natural language that we used to select information will be called "key words".
In addition to the natural language there are other types of language that in contrast with the natural one, are artificial. Among them there are documentary languages, created for information retrieval. Its main characteristic and difference with respect to natural language is that there are no differences between gender and number (they lack of variability), each concept is expressed through a unique term (lack of synonymy) and a unique meaning (lack of polysemes). These terms are called "descriptors".
Natural and documentary languages can be considered like two any other pair of languages. Equivalence between the terms of both can be known using a dictionary. The "dictionary"1 that allows to know the equivalences between the terms of the natural language and those of the documentary language is denominated "thesaurus".
1 Here a "thesaurus" is compared with a dictionary, it must be pointed out that they are not the same. Although a "thesaurus" allows for a translation from the natural to the documentary language, a thesaurus has further elements that make it clearly different from a dictionary.
The thesaurus that allows translating terms from the natural medical language (in English) to the PubMed's documentary language is known as MeSH (Medical Subject Headings).
The procedure for searching is very easy. First of all we must to connect with the data base. Its address is http://www.ncbi.nlm.nih.gov/entrez/query. fcgi?DB= pubmed nevertheless it is not necessary to memorize it, because is enough to write in the navigator either in small or capital letters PUBMED.GOV.
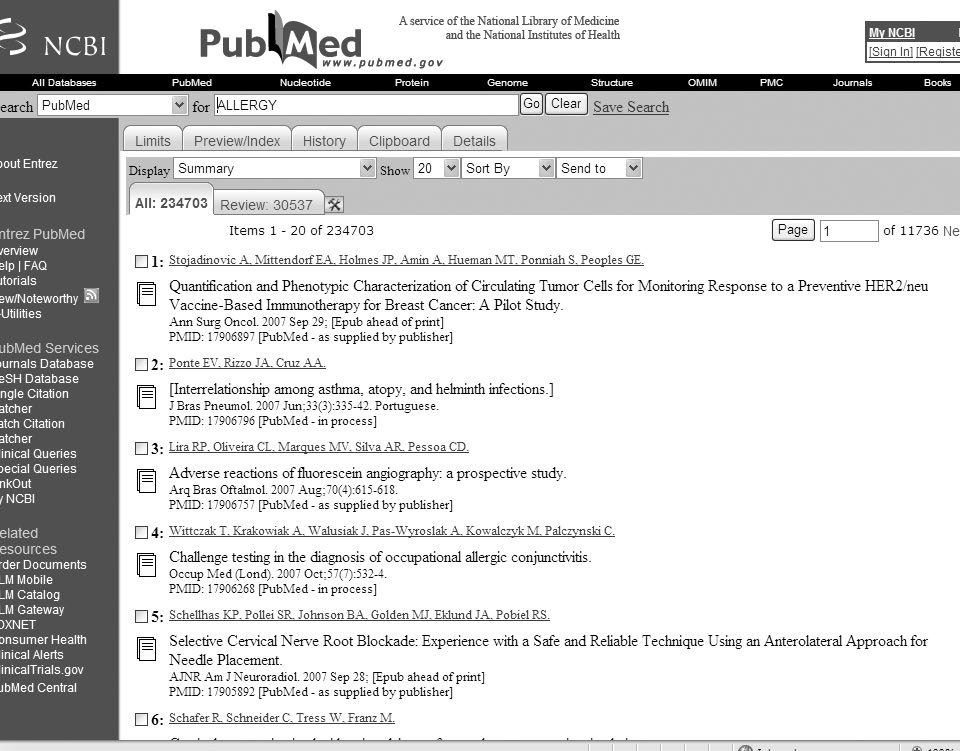
This action will lead to a screen (figure 1 shows this screen with a consultation already done from now it will be used as reference) that has in its upper part a pull-down menu (preceded by the word "Search") that allows the change of database during the search. During the search we must make sure that in that window it appears the word PUBMED.
Figure 1.--PubMed Search Page.
After this menu there is the word "for" and an empty box, the box for consultation, in which the cursor blinks waiting for us to write the term that we have chosen to begin to look for the information. After this box there is the button "go" (that can be clicked with the mouse) although is also possible to use a short cut the key "enter" (↵) of our keyboard that will launch the search, when we have finished typing our consult. The button "clear" will allow us later to erase the complete content of the consultation box. When a search has been done, to the right of the "clear" button it will appear the option Save Search, a utility that requires registration and allows keeping the search done and even allow for an automatic update and the new results are sent to us periodically by e-mail.
The terms that we have chosen can be written in the search box either in capital or small letters. We must remember that we are working in English no special characters like tilde or "ñ" can be used. The letter "ñ" usually is replaced by "n" in the database, in this way "López Piñero" will become "Lopez Pinero" and should be written in that way in the entire search.
TO LOOK FOR THE WORKS OF AN AUTHOR
The simplest search of all that can be done is the one that allows to select all the works in which a certain author has participated.
The procedure is just to type in the search box the last name of the author or authors followed but no necessarily by their initials. Later we must indicate that we look for an author, for this we will include the particle "au" in brackets. With this search we are indicating the software to look for the sequence only in the field that contains the author's names labeled as "au" (table II). For example PINERO A [AU].
This search will deliver as answers all the records which contain in the field Author the word Pinero A, i.e. all the works in which an author named A. Pinero has worked. Nevertheless some rules have to be taken into account:
1.The letter "ñ" must be changed by "n" written accents like "á", "é" etc., can not be used.
2.The first initial of a name includes all the compound names. In the example if we have selected Pinero A, that would have included also AM, AC, AL, etc. If we only want to select those works from AC Pinero, our search should have been: PINERO AC [AU]2.
2 From now on, the text that must be typed in the search box will appear always in capital letters although (with an exception that will be studied later) capital or small letters can be used indistinct.
3.Last name doesn't include double last name. In the example search all authors whose first name begins by A and that have only Pinero as last name. The search excludes those works whose author is PINERO FERNANDEZ3, PINERO MARTINEZ, PINERO SANCHEZ or LOPEZ PINERO. To find all the papers whose author is José María López Piñero, the search should be LOPEZ PINERO JM [AU].
3 From now on the words typed in the search box will follow the database rule even if that means as in this example that the Spanish orthographic rules are overcome as in this example when accents were omitted.
4.It is not necessary (although it can be done) to join both last names with a hyphen. The search for LOPEZ PINERO [AU] will deliver the same results than LOPEZ-PINERO [AU].
BIBLIOGRAPHIC SEARCH IN NATURAL LANGUAGE
Usually the most common situation is not to search for an author's papers but to search in a certain thematic area which concerns us when we perform the search.
The procedure mentioned above is to select those terms related with the problem that concern us. E.g., let us think that you are interested in some problem related with allergy. It seems logic that one of the key words in the search should be "Allergy". In order to select the papers we will type ALLERGY in the search box and will launch the search clicking the button go. As result, the last line of the heading of the search page will inform us of the number of records obtained with a sentence like this "Items 1-20 of 234684". That means that when the search was done, 234684 records were found and 20 items per page are shown. Below this information begins the display of the records founded, ordered by chronologic order from the newest to the older.
Over this results line, additional information is offered. The tab "All:" allows the selection of all the records whose profile agrees with the search launched. The Tab "Review:" shows a selection of the same documents after an additional filter, that can be created clicking on the button that is located to the right of the tab and leads to "My NCBI" (it can also be accessed from the blue column in the left of the screen); this is a service that require registration for an account. The software will keep in its memory the filters that we used (till a maximum of 5) and will be used systematically in all searches that we will do. This service is very useful when we must do repetitive search with specials requirements (such as language, or type of document)
It seems evident that the search that we have just done in the example is not very useful because it is unspecific, but it is going to help us to understand another of the characteristics of PubMed. Un like other databases, this does not merely search for a certain string typed letters (in "A-L-L-E-R-G-Y" in our example), but is programmed to offer us as answer the maximum amount of information and "interprets" which is the meaning of our search. In order to know what the interpretation that PubMed has done is we must push on the "Details" tabs. This action will lead to a new screen where we will be informed that our request for information (it has been the word "ALLERGY") typed in the search box has been interpreted as
"immediate hypersensitivity"[Text Word] OR "hypersensitivity, immediate"[MeSH Terms] OR ("hypersensitivity"[TIAB] NOT Medline[SB]) OR "hypersensitivity"[MeSH Terms] OR ("allergy and immunology"[TIAB] NOT Medline[SB]) OR "allergy and immunology"[MeSH Terms] OR Allergy[Text Word]4
4From now on, in the search operators such as OR or AND will be used. In a later section they will be studied in detail.
In contradistinction to other databases, the software has introduced new terms (hypersensitivity, immunology) that were not included in the original search allowing increasing the sensibility of the search. The explanation is not that we are working with artificial intelligent software but a special mode of search known as Automatic Term Mapping).
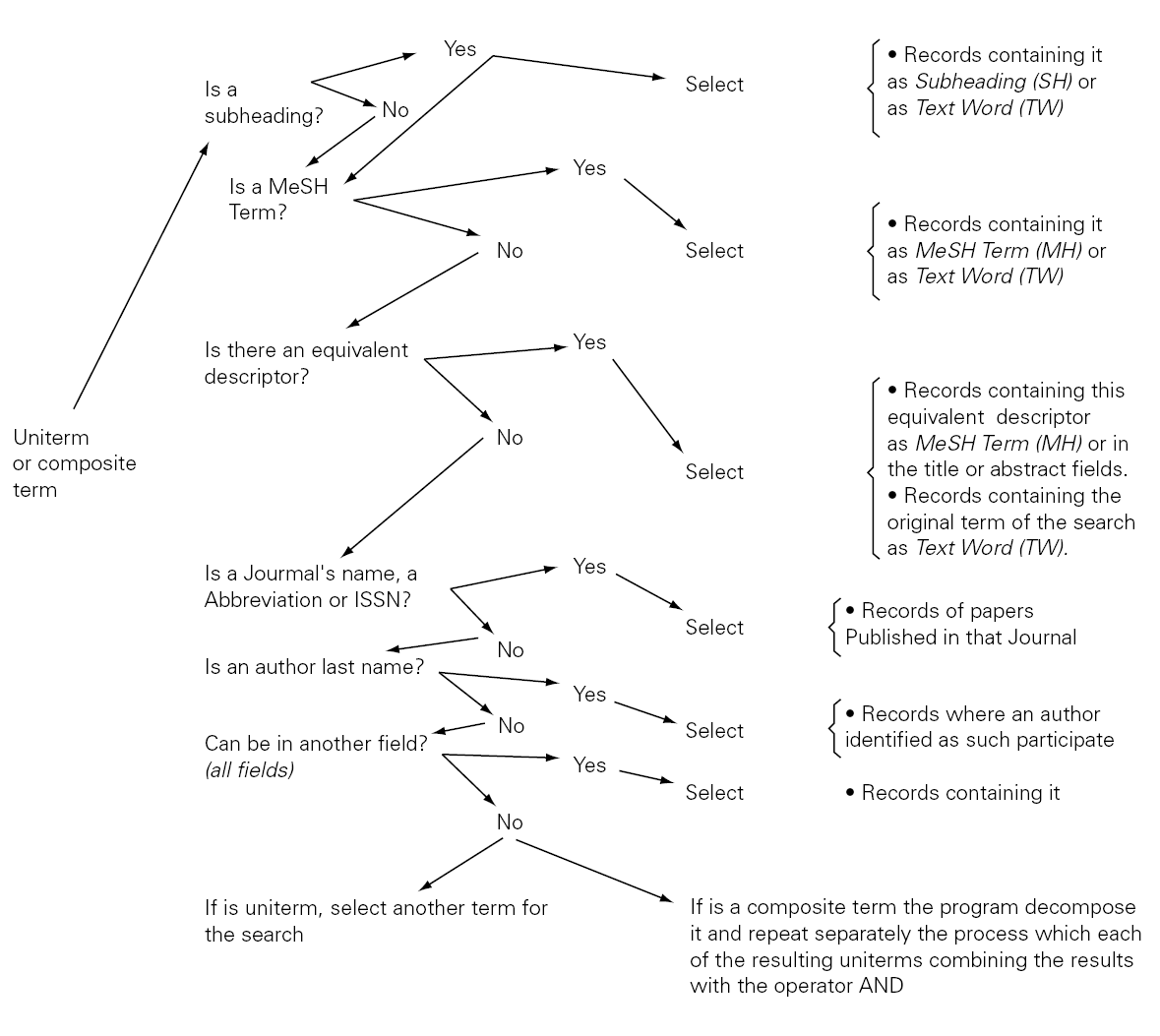
AUTOMATIC TERM MAPPING
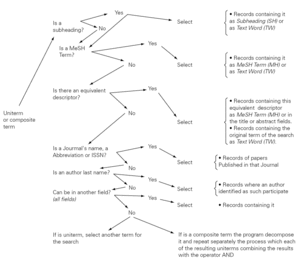
The Automatic Term Mapping (ATM) is a system designed to provide a high sensibility in information searches, without needing that the user has a special training... The launching of the search order unleashes the search of the string in one field after other in each of the register of the database. It the string is found in the first field the search will stop, if not it will go on in a different field. A schema of the ATM is presented in figure 2.
Figure 2.--Automatic Term Mapping.
The schema shows that the search can begin in any uniterm (as the one that we have been using ALLERGY) or a composite term (e.g. IMMEDIATE HYPERSENSITIVITY). When the search order is launched the software will take all the string of characters (including spaces) and will carry out all the steps described in figure 1 till it finds an answer that can be given to the user.
The ATM intention is to provide the maximum amount of information (a high sensibility in the search) as well as a relatively high specificity, although sacrificing part of the last one. That often leads to a high level of "noises". Furthermore, in some occasions, the answers are not adjusted to the user demands. Due to that some times may be convenient to cancel the execution of ATM, that can be done by the user itself orienting the search to certain fields or specific record subgroups, as well as using the truncate term or enclosing it in quotation marks, as it will explained later.
INFORMATION SEARCH IN CERTAIN FIELDS OR SPECIFIC RECORD SUBGROUPS
For those that have enough experience using PubMed is easy to predict which will the translation of the search request and using it to obtain high quality information. Nevertheless some time, is desirable to cancel the ATM that can be done in several ways. The first one is to enclose the search term in quotation marks that is equivalent to ask the program to search exactly this string in any of the fields of each record ([All Fields]). For example, the search "ALLERGY", in quotations mark, doesn't launch the ATM, but selects all the records that contain that word in any of the fields ([All Fields])
The second way of deactivating the ATM is to indicate the software in which field it must search the string that have been typed in the search box, that can be done adding between square brackets the label that identifies the field (see table II). For example the above search where we wrote ALLERGY without indicating anything else, changes dramatically when the following transformation are used.
User's Order: ALLERGY [AD]
Software's translation: Records that contain the string ALLERGY in the author address, therefore, these are papers written in institutions or organizations consecrated to allergy.
User's Order: ALLERGY [TI]
Software's translation: Records that contain the string ALLERGY in the English title and therefore its content may be related with allergy.
The possible variations are as much as fields exist in the records, the information obtained is wholly different as it can be seen. Table II shows a set of fields that are more often used in search and that will allow us for example to select works that have been published either in certain dates or countries or by certain authors or institutions.
Some times, depending of the user's information needs especially if these are the solving of a clinical problem it is very interesting to look for some types of publications such as clinical practice guidelines, clinical trials, systematic reviews, meta-analysis etc. As one of the field is publication type, the search can be oriented specifically to select on of these, that can be achieved adding to the term that defines the search the string AND°$[PT], where°$ is the type of publication that we want to obtain. (The whole list appears in Pub Med's Help-http://www. ncbi.nlm. nih.gov/books/bv.fcgi?rid=helppubmed. table.pubmedhelp.T42-). For example, if we would like to look for randomized clinical trials on anti-allergic drugs that have been Published, one of the options for searching for information (not the only one) could be:
"ANTIALLERGIC AGENTS" AND CONTROLLED CLINICAL TRIAL [PT]
The software's answer will be to offer the list of all the records of controlled clinical trials that contain the string ANTIALLERGIC AGENTS in any of its fields (due to the quotation marks that cancel the ATM execution).
In the same way that the search can be oriented toward a specific field, it can be also oriented toward a specific subgroup of record among those presented in table I. To limit the selection in one of the groups classified by its content or by its location in the record AND LLL[SB] must be added to the search, where LLL the denomination of the subgroup in the table. For targeting the search to a set of records from specialized journal, the sentence AND JSUBSET*** should be added, where *** is the letter or group of letters that identifies the group in table I.
USE OF BOOLEAN OPERATORS5 FOR SEARCHING INFORMATION
5George Boole (UK, 1815-Ireland 1864) was an autodidact mathematician. He wrote 50 papers and books and was one of the first that studied numbers proprieties. He worked also in differential equations (his influent Treatise on Differential Equations was published in 1859), in the calculation of finite differences (Treatise on the Calculus of Finite Differences, 1860) and on the general methods of probability. In 1854 he published Investigation of the Laws of though, a classical milestone in the history of mathematics, that is the origin of the Sets theory and part of the mathematical theories on logic and probability. Boole formulated logic as algebra, pointing out the closeness of algebraic operation with logic operations. His algebra consists of a method for solving logic problems, he uses binary value 1 and 0 (that represents opposed pairs present/absent, true/false) and the logic operations AND (and), OR (or) and NOT (no). He initiated Boolean algebra that became the foundation of practical digital circuit and provided the theoretical grounding for the Digital Age as well for the management of information in data bases.
Very often, a unique term (either uniterm or a composite term) is not enough to define our field of interest and it is necessary to use more that one term. If we are concern about the use of antihistaminic H1 in allergy, it is obvious that the terms H1 ANTIHISTAMINICS or ALLERGY, individually, are too generic to precisely define our interest. Somehow we should be able to use both terms simultaneously. As happen in all the languages those terms must be linked conveniently to construct sentences that make sense.
To link one term with another the Boolean operators AND, OR and NOT are used. Although the search terms may be typed indistinctly in capital or small letters, Boolean operators must always be in capital letters.
OR: Operator OR is an inclusion operators. A search of the sort "A OR B", will select all the records that comply either with the A requirement, the B or both of them. It is possible to link terms with the operator OR so a sentence like "A OR B OR C OR D OR E OR...", that will select all the record that comply with one of the requirement A, B, C, D, E..., can be built.
A special case related with operator OR is that of truncated terms that is also a way of avoid those problems due to the variability of natural language. Many terms are made bay a common root and ending that modifies its meaning. (e.g. ALLERG- EN /ENS /IC /ISTS /Y... etc). If a selection that include all these terms is wanted, all these terms should be written joined by the operator OR; there is a simple and easy way, to write only the root followed by an asterisk (i.e., ALLERG*), it will select all records that contain any term with the root ALLERG-. Truncation must be used with precaution due to two reasons, first the same truncated terms may refer to different concepts and not related between them. (One extreme example: the search HYPER* can be done but it don't make any sense); this danger increases as the root becomes shorter; it is recommended that it shouldn't have less than five letters. The second reason is that when the root diversifies in a great dealt of different terms, the software doesn't select all, but a limited number of variations, the first ones in alphabetic order. That is the case of the truncated term ALLERG*, that won't provide adequate results. Furthermore the truncation of a term also cancels the ATM, as was mentioned above.
AND: The introduction of the operator AND between two terms A, B will select all the records that fulfill requirement A and that in addition fulfill requirement B. It is also possible to link terms and operator in one sentence like A AND B AND C AND... The selection will show all the records that fulfill each of all the requirements A, B, C...
NOT: Operator NOT is an exclusion operator. The inclusion of a NOT between terms A and B, will select all the records that fulfill requirement A, except those that fulfill B. As in the above operators it is possible to create a sentence such as A NOT B NOT C NOT... that will select all the records that comply with A requirement except those who comply with any of the following (B, C...).
It is important to give attention to the order in which the terms of the search and the operators are written because as any language the meaning of the sentence changes and therefore the results obtained in the search. Although A OR B is equivalent to B OR A, and A AND B is equivalent to B AND A, A NOT B is the oppose to B NOT A
Furthermore the same search may combine different operators, so it could be possible a sentence like A OR B* AND C NOT D... By defect, the program, will execute instructions from the left to the right, i.e. in this case:
1.It will select all the records that comply with A,
2.It will add those with comply with B* (i.e. those that contain a Word with the root B, whatever it is).
3.From these it will only select those that in addition comply with C.
4.Last those with comply with D will be excluded
Nevertheless instructions can be changed by the user either varying the terms and operators order or including brackets in the sentence, in that way the search A OR ((B* AND C) NOT D), would be equivalent to B* AND C NOT D OR A, i.e.:
1.Select all the records that comply with B*,
2.From these, select only those with in addition comply with C,
3.From the remaining exclude those with comply with D,
4.To the remaining group it will add all those with comply with A, no matter they comply or not with B*, C or D.
It is necessary to take into account that the program will execute the implicit order in the operators after having working with the individual instructions applied to the terms, so operator will act after ATM have been executed, or if a instruction has been given, after the selection in the chosen field. E.g. A OR "B" AND C[TI] NOT D[LA]
1.Select all the records obtained from applying ATM with term A,
2.Add those that include term B in any of the fields (all fields),
3.From the records select only those that in addition contain C in the English title.
4.Last exclude from this selection those published in language D.
With a little of practice is possible to build phrases with a very precise sense combining operator and field delimitations (tags).
PUTTING LIMITS TO INFORMATION
Once that the terms to be used in the search, the fields to be selected have been decided and how these terms will linked to build a sentence that make sense, the search can be limited with certain criterions.
This can be achieved if, in the search page, we select the tab "Limits". This action allows to reach a series of popup menus (fig. 6) that can be used to select the field toward the search will be oriented, the type of publication that will be obtained, the language in which has been published, the related age group, if it is an experiment either in humans, animals or both, in men or in women, date of publication... The action is equivalent to introduce between brackets the search that we have designed and to add several AND followed by the choice and the appropriated field.
An example: In theory papers on the pharmacologic treatment of allergy can be obtained, among other searches, with the following: "ALLERGY" AND "DRUG THERAPY". Limits to this search can be added choosing in "Limits" the type of publication (e.g. Randomized Controlled Trial), the language (e.g. English), the age (e.g. adolescent) and the date of publication (e.g. from 1995 to 2004)
It must be taken into account that the chosen limits will apply to the entire search. All of this will be translated by the program to:
"ALLERGY"[All Fields] AND "DRUG THERAPY" [All Fields] AND Randomized Controlled Trial[ptyp] AND Spanish[Lang] AND "adolescent"[MeSH Terms] AND ("1995"[PDAT]: "2004"[PDAT]).
The results screen will show under the tab "Limits" the limits that we are applying to the search.
This program translation is equivalent to directly typing in the search box the following
"ALLERGY" AND "DRUG THERAPY" AND RANDOMIZED CONTROLLED TRIAL[PT] AND SPANISH [LA] AND ADOLESCENT [MH] AND 1995:2004[DP]
However, this second procedure has advantages on the use of the "Limits" menus because it allows for more options and to apply different limits to different terms in the search.
USE OF THE INDEX IN THE INFORMATION SELECTION
In the search screen under the search box, the tab "Preview/Index" provides an option that can be comfortable in the search of information although it has limitations.
The Index is only a list of all the terms (uniterms o composite) with a meaning that are included in the Data base. This option allows selecting in the popup menu on the second half of the screen a field where to look for the term that will be typed in the following line. The selection of the button Index will show us all the uniterm and composite terms that contain the typed word.
It is possible in this list, holding the CTRL, key to select all those that we consider appropriated, to be combined using operator OR. After that we will choose the operator that we want to use to link the selected terms with the active search that we have, the linked term will be sent to the search box and the buttons Preview o Go will launch the search. Preview will add the search to Most Recent Queries area and it will indicate the number of obtained records. Go, will take us back to the search page and will show as the list of the selected records.
SEARCHES WITH THE MESH THESAURUS
Till now we have known the Basic possibilities of information search using only natural language. I.e. in a systematic way we have use the terms (key word) that we thought were more adequate, without considering if these were the standardized terms that make the documental language used by the program (descriptors or in the program language MeSH Terms).
The alternative is to work directly with descriptors. The final results of this a less exhaustive selection but more precise. The first thing to do in this kind of searches is to know the descriptor equivalent to the key word that we have chosen.
For doing this we must access to the MeSH thesaurus, we can do in two ways from the search screen: selecting MeSH in the popup menu Search, o choosing MeSH Database in the blue column in the left side of the screen.
Whatever option is chosen will lead us to a new screen where the cursor blinks in the search box and below explains us that MeSH is the NLM controlled vocabulary used for indexing papers for MEDLINE/ PubMed that provide an efficacy way of retrieving information without using different terms for the same concepts.
From this can be drawn as was mentioned earlier, that the MeSH thesaurus is in certain way a "dictionary" that allows us to translate from the natural language (in English) to the descriptors (also in English) avoiding in the searches problems with language variability, synonymies and polysemes.
The first step will be to translate a selected key Word to its equivalent descriptor, that will be done typing in the search box (alter checking that the popup menu Search shows the option MeSH) and launching the search by pressing Go.
Lets assume that we are in a clinical situation that ask us to solve a certain problem in a patient with suffers from allergy. What is the adequate descriptor to search for information on allergy? If we have this keyword "allergy" we will type in English in the search box: ALLERGY.
Whatever search we do the answer will be similar. Under the heading and after the term Suggestions a set of terms either conceptually or orthographically similar to that used by us will be offered, that allows pressing on them to change from our departure search.
Below will appears a listing of those related with the chosen key word followed, when it is necessary by its definition. If the chosen keyword is at the same time a descriptor, it will appear on the first time, followed in occasions by other descriptors. If the keyword is not a descriptor the list will be headed by another different descriptor. The last one is the case if we have used the term ALLERGY.
The following step is to select from those offered by the program, and assisted by the accompanying definition the closest descriptor to the concept that we are looking for, we will do that pressing over it. For example the keyword ALLERGY drive us among many, to the Hypersensitivity, Immediate. If we select this (or other that adjust betters to our expectative) it will led us to a new screen where the selected descriptor will appear as well as its definition when it's adequate.
Now under the descriptor, all the subheadings that have allocated to it will appear. We will be able to decide just by parking the pertinent boxes, what concrete aspects interest us. If we don't check anyone, we should check the box that precedes the descriptor and the selection will comprehend all the subheadings.
After the subheadings, the option Restrict Search to Major Topic headings only, allows us by selecting it, limit the search to those papers in which the descriptor represent the main topic of the paper. In case if not selected we will obtain all the papers that deal with the topic even in a secondary way.
Entry Terms indicates the key words that have been programmed as the descriptor's synonyms. They are keywords that lead to the descriptor. Under they the descriptor is presented inserted in one or more generic-specific trees, where is related with other members of its semantic family sorted according the amplitude of its meaning from those of the wide meaning (toward the upper left) to those with an stricter and concrete sense (toward the lower right).
Lets assume that in the search that we have been doing in this section we have selected the descriptor Hypersensitivity, Immediate. The generic-specific tree will show us in the upper left of this term the descriptor Hypersensitivity indicating that this is a more generic meaning and includes all that is located below (i.e. Hypersensitivity, Immediate is a part of Hypersensitivity). Below to the right of Hypersensitivity, Immediate there are other terms with a more restricted meaning including in the above (i.e., "Hypersensitivity, Immediate" includes "Anaphylaxis", "Conjunctivitis, allergic", "Dermatitis, atopic", etc.) and those are in other more restricted (e.g. "Food hypersensitivity" including hypersensitivity to eggs, milk ...). The symbol + that follows to some of the descriptors indicates that this can be subdivided in other more specifics.
If we launch the search from this point, the selection will include not only the records to be allocated to the active descriptors. (In our case Hypersensitivity, Immediate), but also all the descriptors with a more restricted meaning ("Anaphylaxis", "Conjunctivitis, allergic", "Dermatitis, atopic"...). To limit the search to the only active descriptor the option Do Not Explode this term must be used.
The generic-specific tree allows us a final decision: to change or not to change the descriptor choice. Let's assume, that in this point, Hypersensitivity, Immediate, seems to us, in view of its location in the tree a term too generic for our original intention. We can change into any of the descriptors, e.g. Anaphylaxis, by clicking on it, what leads to a similar screen where again we can choose (or not) the subheading that we are interested, to decide if we wish only those papers where the new descriptors is the main topic, if we want that the selection include or not more specific descriptors or even to change again the descriptor.
When all the decisions, have been taken, the popup menu Send to allows us to send the selection to a search box where it appears the translation of the sentence that we built that the program does. There are three options for Search box that allows adding new terms to the box, combining it with the terms already covered through the selected operator. Finally if we agree with the program's translation of the phrase that we have built, Search PubMed will give as answer the list of all the papers that fulfil the search requirement.
UTILIZATION OF THE SEARCH MEMORY
OR HISTORY
As we develop our work PubMed is keeping a file on all the searches we do.
We may see it, if in the PubMed search and under the search box, we select the tab History. This leads to a new screen where it will appear ordered all the searches that we have made during the session.
Each query is numbered and the number preceded by the symbol #. Following the search the number of records obtained in the query is shown. From History we can carry out, among others, two actions: Go back to an earlier query, and to combine different search with the Boolean operators that we decide.
To return to an earlier query without having to retype again it is enough to click on the figure that indicates the number of records retrieved.
To combine several searches will write in the search box the number that identifies preceded by # and we will connect them with the operators that we consider necessary. E.g. P.e. #1 OR #4 AND #5 NOT #7. It should remembered that here are also applicable the standards specified above for consultation with Boolean operators. Also by clicking on the menu which identifies the search will pop up a menu allowing sent to the search box the operator chosen (AND, OR, NOT), remove the search from the memory (Delete), to return to the search (Go), return to the translation done by the program (Details), or if we are registered save the query into a personal space. (Save in my NCBI).
WIDEN THE SELECTION: RELATED ARTICLES
When we arrived with our quest to a paper that appeared tailored to our information needs, we can expand the selection to similar documents by simply selecting Related Articles to the right of the chosen record.
This causes the program to make a new selection, which will include documents that share certain characteristics, and which has served as a starting point, essentially descriptors and keywords. Furthermore PubMed will offer this selection ordered by its proximity to the content of the initial document.
CLINICAL QUERIES
One of the services offered by PubMed with the name of Clinical Queries it is very interesting because it direct application to clinical work. It is located in the blue column at the left of the search screen.
Aside from the Medical Genetics Searches option that is not related with the objectives of this paper, Clinical Queries allows us two alternatives. In one, Find Systematic Reviews based in the subject that we have selected and written in the search box, we will get systematic reviews and meta-analysis on it.
In fact the computer program does the selection not throughout the whole database, but in a special group that we have already known, that of systematic reviews (see table I). The results are the same that we will get in the normal search box if we add to the final strategy AND SYSTEMATIC [SB].
The second option, Search by Clinical Study Category, allows the selection of clinical oriented papers. The method is similar: we must specify in the search box the area that we are interested in. After that we will choose among one of the five possible categories (treatment, diagnosis, etiology, prognosis or clinical prediction guides) that fits to our needs. Later we will have to choose between obtaining a wide selection, including the most important papers but perhaps also some noise (sensitivity), or a narrower range that includes only the most relevant, but that can leave out some interesting papers. (specificity).
When we launch the search, the program will automatically apply to the selection a set of filters developed by Haynes RB et al in a classic work that can be consulted at the following address http://www.nlm.nih.gov/pubs/techbull/jf04/ cq_info.html
An example. If someone it is interested in the pharmacologic treatment of anaphylaxis, we already have explained how to use the thesaurus. Through it we could arrive among other things to the expression "ANAPHYLAXIS/DRUG THERAPY" [MH] (ie. Papers that contain the anaphylaxis descriptor and pharmacologic treatment drug therapy- as subheading). This heading will give us a wide selection that we can limits. (Remember the use of Limits, operators and field delimitations) according to our interests. Let us suppose, for example, that we only want papers in English our sear would look like: "ANAPHYLAXIS/DRUG THERAPY"[MH] AND ENGLISH[LA]
At this point you can select among these papers only systematic reviews and meta-analysis. To do so, ad at the end of the search AND SYSTEMATIC[SB], or select and copy the strategy, take it to Clinical Queries, select Find Systematic Reviews, paste the clipboard on the search box and launch the search.
If we are interested in clinical oriented papers, we can begin from Clinical Queries, for example with the expression ANAPHYLAXIS[MH] AND ENGLISH[LA] in the search box Search by Clinical Study Category. Besides we will select the option that we are interested in Category (e.g. therapy) and the emphasis on Scope (sensitive or specific).
SEPARATE RESULTS TO THE CLIPBOARD
When a selection is done, in many cases we are no interested in saving all the records, but only some of them. In each of these queries we can separate flagging it the records that we want up to a maximum of 500.
Once marked in the popup menu Send to we select Clipboard and the records will be stored in the clipboard. To view it contents we must select the tab Clipboard that appears under the search box.
Once inside the clipboard we can undo any selection by marking the record and selecting Clip Remove from the Send to menu.
Correspondence:
J.M. Sáez Gómez
Facultad de Medicina. Campus de Espinardo
30100 Murcia. Spain
Phone: + 34 968-367179
Fax: + 34 968-36 4150
E-mail: jmsaez@um.es
BIBLIOGRAPHY
In these pages is not possible to analyze all the aspects and possibilities of PubMed, furthermore it is updated and modified very often. Due to that printed papers on the use of PubMed become usually outdated and it is better to collect information from the web, nevertheless we should apply a logic filter: select only those places that are updated regularly. Additional information can be obtained from:
PubMed Overview (http://www.ncbi.nlm.nih. gov/entrez/query/static/overview.html) provides a good introduction on PubMed and its characteristics.
PubMed Help (http://www.ncbi.nlm.nih.gov/ books/bv.fcgi?rid = helppubmed.chapter.pubmedhelp) this is the PubMed Help. Very complete and clear. It provides information about all the aspects mentioned in this paper and something else.
PubMed Tutorial (http://www.nlm.nih.gov/bsd/ disted/pubmedtutorial/) an excellent tutorial with animated demonstration on the use of PubMed.
The above three sites depends of the own National Library of Medicine, they are accessible from the database, they are updated when ever is necessary. Very recommendable.
Buscar en Medline con PubMed (http://www. fisterra.com/recursos_web/no_explor/pubmed.asp) A tutorial provided by the portal Fisterra. Very good as a first contact with PubMed, it has also the advantage of been in Spanish.
Tutorial para la búsqueda en PubMed (http:// www.sap.org.ar/staticfiles/medline/tutorial/index.htm) Interactive tutorial made by the Health Science Center Library from the University of Florida, translated into Spanish by the Sociedad Argentina de Pediatría.
DECS-DESCRIPTORES EN CIENCIAS DE LA SALUD
(http://decs.bvs.br/E/homepagee.htm) This can be considered in certain way a Spanish version of the MeSH-Medical Subject Headings de la U.S. National Library of Medicine. It was developed from it with the aim of allowing the use of a common terminology for searching in three languages (Spanish, English, and Portuguese), providing a consistent and unique environment for retrieving independently of the language. In addition to the original MeSH terms, specific areas for Public Health and Homeopathy were developed
IMPROVED SEARCH IN PUBMED
(http://www.infodoctor.org/rafabravo/pubmedes.html) For quick and easy searches in PubMed.