Lipid transfer protein (LTP) is a major fruit allergen. It has, however, recently been revealed that the systemic reaction in peach-allergic patients is related not only to LTP (Pru p 3) but also to gibberellin-regulated protein (Pru p 7). We investigated recombinant Pru p 7 (rPru p 7) for its potential use in worldwide standardization for the diagnosis of peach allergy.
MethodsNatural Pru p 7 (nPru p 7) was purified from peach crude extract using a monoclonal antibody affinity column. Complementary DNA for Pru p 7 was cloned and expressed in Escherichia coli and Pichia pastoris. Serum immunoglobulin (Ig) E in peach-allergic patients was examined by enzyme-linked immunosorbent assay (ELISA) using nPru p 7 and rPru p 7 (E. coli product: erPru p 7 and P. pastoris product: prPru p 7).
ResultsPeach-allergic patients (n=27) were diagnosed and categorized into oral reaction (n=10) or systemic reaction (n=17). The nPru p 7 positivity based on serum IgE levels was 52% in the systemic-reaction group and 0% in the oral-reaction group (P<0.05). In the systemic-reaction group, there was no significant difference in reactivity between nPru p 7 and prPru p 7, but the reactivity of erPru p 7 was significantly lower than those of nPru p 7 and prPru p 7 (P<0.05).
ConclusionsWe found that prPru p 7 exhibited reactivity in ELISA comparable to that of nPru p 7 for the diagnosis of peach allergy with systemic reaction.
Peach (Prunus persica) allergy is one of the most common allergies in the Mediterranean region and has been reported in Japan.1–7 The number of reports of peach allergy concerning component-resolved diagnosis (CRD) has recently increased.8–10 CRD may be useful for clinical-type classification.6,11 Although Pru p 3 is known to be a popular plant allergen component, some cases of systemic reaction (SR) cannot be explained by Pru p 3.6,12,13 Pru p 3 is typically present in the outer part of the peach, and it sometimes does not cause symptoms when the peeled peach is eaten.14 Tuppo reported Pru p 7 as a new peach allergen in Europe.15 In Japan, many peach-allergic patients with SRs have shown sensitivity to Pru p 7, although few patients have shown sensitivity to Pru p 3.6,16 Therefore, the contribution of Pru p 7 to peach allergy should be revealed promptly and globally. However, it is very difficult to obtain Pru p 7 free from Pru p 3 because of their similar protein chemical properties.15,17 Therefore, the development of recombinant Pru p 7 (rPru p 7) is an urgent issue for the advancement of Pru p 7 research and diagnosis.7,18,19 For the expression of recombinant proteins, P. pastoris is widely used.20–25 Pokaj found that P. pastoris is superior to Escherichia coli as an expression system for the production of large quantities of soluble, properly folded, and biologically active rCor a 8 [hazelnut lipid transfer protein (LTP)].26
In this study, we comparatively examined serum immunoglobulin (Ig) E levels in peach-allergic patients by enzyme-linked immunosorbent assay (ELISA) using natural Pru p 7 (nPru p 7) and rPru p 7 (E. coli product: erPru p 7 and P. pastoris product: prPru p 7) purified using monoclonal antibody (mAb) columns.
MethodsStudy participantsA total of 84 participants with fruit allergies (including allergies except peaches) were enrolled in the Fruits Allergy Component Study Group (http://www.fruit-allergy.jp/) from June 2014 to December 2015. After enrolment, we applied to the ethical review boards of each facility and examined only those who were approved after obtaining written informed consent. Exclusion criteria included participants who had no symptoms when they ate peach or those whose symptoms were unclear or not reproducible. After obtaining informed consent, the participants answered the questionnaire and then underwent a skin-prick test with peach. This study was conducted according to the World Medical Association's Declaration of Helsinki. Ethical approval was obtained from the Fujita Health University Ethics Committee in July 2014 (reference number: 14-075) for all sites taking part in this study.
Definition of symptomsWe evaluated the following types of symptoms induced by the ingestion of peach according to the participant's questionnaire as follows: Oral reaction (OR): Itching or tingling sensations in the oral mucosa, palate, or throat that developed within 5–10min after peach ingestion and localized symptoms with no SR.27 SR: Symptoms presented in ≥2 organs after peach ingestion. Systemic symptoms included anaphylaxis and food-dependent exercise-induced anaphylaxis.
Preparation of peach crude extractPeaches (P. persica, cultivar Asama–Hakutou strain) at the commercial ripening stage were obtained from a local store. Entire peach fruits (peel and pulp) were homogenized with an extraction solution (2mM disodium ethylenediamine tetraacetate, 10mM sodium N,N-diethyldithiocarbamate, 3mM sodium azide, and 2% solid polyvinyl polypyrrolidone) at a 1:1 [w:v] ratio. After filtration through gauze, the homogenate was centrifuged at 10,000×g for 15min at 4°C. A cation exchange resin (Toyopearl CM-650M; TOSO, Tokyo, Japan) was added to the supernatant and mixed overnight at 4°C. The resin was collected by centrifugation, packed, and washed with 20mM phosphate buffer at pH 5.0 in a column. The proteins absorbed to the resin were eluted as the crude extract with 0.5M sodium chloride in the same buffer.
Production of mAbs and purification of natural antigensThe production of hybridomas producing mAbs specific to peach Pru p 7 or Pru p 3 and the purification of natural antigens were essentially as described previously.28 The hybridomas that produced IgG reactive to the crude extract were cloned twice with the limiting dilution method and inoculated into pristine-primed mice. Then, mAbs were prepared from the ascitic fluids and purified on Protein G-Sepharose columns (GE Healthcare, Fairfield, Connecticut, USA). The mAbs specific to peach Pru p 7 or Pru p 3 were selected using an immunoblotting analysis with the crude extract under non-reducing conditions. The animal experiments were performed under the guidelines of the Animal Experiment Committee of Kyoto Women's University following a bulletin (No. 71, 2006) from the Ministry of Education, Culture, Sports, Science, and Technology in Japan.
Pru p 7 or Pru p 3 was purified from the crude extract using a HiTrap NHS-activated HP column coupled with each mAb according to the manufacturer's instructions (GE Healthcare).
Creation of recombinant antigensTotal RNA was isolated from peach from 200 to 400mg net weight using Sepasol (Nacalai Tesque Inc., Kyoto, Japan) according to the manufacturer's protocol. Complementary DNA (cDNA) was obtained using 0.5μg total RNA, 100U reverse transcriptase II (Invitrogen, Carlsbad, CA, USA), a random primer, 10mM dithiothreitol (DTT), and 20U RNase inhibitor (Nacalai Tesque Inc.) in the presence of a reaction buffer provided from the manufacturer. The full-length Pru p 7 (locus name ppa014086 m.g, Phytozome site, https://phytozome.jgi.doe.g.,ov/pz/portal.html) was obtained by polymerase chain reaction (PCR) amplification. The primers used were 5′-GGTTCATCTTTCTGTGACTCCAAGT-3′ and 5′-TTAAGGGCATTTGGGGTTGCCCTTG-3′. Restriction endonuclease sites were extended at the 5′ end of both primers. The PCR reaction was performed through 25 cycles at 94°C for denaturing, 55°C for annealing, and 68°C for extension. The amplified product was visualized using agarose electrophoresis and ethidium bromide staining. First, the product was cloned into pZERO2 (Invitrogen), and the correct construction of the plasmids was confirmed by sequencing. The Pru p 7 cDNA was digested using BamHI and XhoI and cloned into the BamHI and XhoI sites of pGEX-6p-2 (GE Healthcare) to make pGEX-Pru p 7 for the expression in E. coli and cloned into the SnaBI site of pPIC9K (Invitrogen) to make pPIC9K-Pru p 7 for the expression and secretion into the medium of P. pastoris strain GS115 (Invitrogen).
To induce the expression of GST Pru p 7, 0.2mM IPTG was added in the Luria–Bertani media/50μg/mL ampicillin and incubated at 30°C for 4h. E. coli was pelleted by centrifugation at 10,000×g for 8min and was suspended in PBS at pH 7.4 containing 0.1mM DTT and 0.2mM phenylmethylsulfonyl fluoride. After sonication four times at a power of 12W×30s each time, the clear lysate was obtained by centrifugation at 10,000×g for 30min. After the addition of glutathione beads to the lysate and overnight incubation, erPru p 7 was released from the beads by treatment with PreScission Protease according to the manufacturer's instructions (GE Healthcare).
For the expression of prPru p 7, pPIC9K-Pru p 7 was transformed into P. pastoris. Land transformants that did not require histidine were selected and expanded in buffered minimal glycerol medium (100mM potassium phosphate, pH 6.0, 1.34% Yeast Nitrogen Base, 4×10−5% biotin, and 1% glycerol). The induction of prPru p 7 was achieved using buffered minimal methanol medium (100mM potassium phosphate, pH 6.0, 1.34% Yeast Nitrogen Base, 4×10−5% biotin, 0.5% methanol) at 30°C, and the expression of prPru p 7 was examined in the culture supernatant.
ErPru p 7 and prPru p 7 were purified using HiTrap NHS-activated HP columns coupled with anti-Pru p 7 mAb similar to the purification of nPru p 7.
Protein analysesSodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed at a constant current of 80mA under reducing or non-reducing conditions. The gel was dyed using Coomassie Brilliant Blue R-250.
The proteins on the gel were transferred to a PVDF membrane (BIO-RAD, Hercules, California, USA), and the membrane was blocked with PBS containing 5% skim milk for 1h at room temperature. After incubation of the membrane with anti-Pru p 7 or Pru p 3 mAb (hybridoma culture supernatant) for 1h at room temperature, alkaline phosphatase-labeled anti-mouse IgG (American Qualex, San Clemente, CA, USA) was added and incubated for 1h at room temperature. The color reaction was performed using an alkaline phosphatase coloring kit (Nacalai Tesque Inc.).
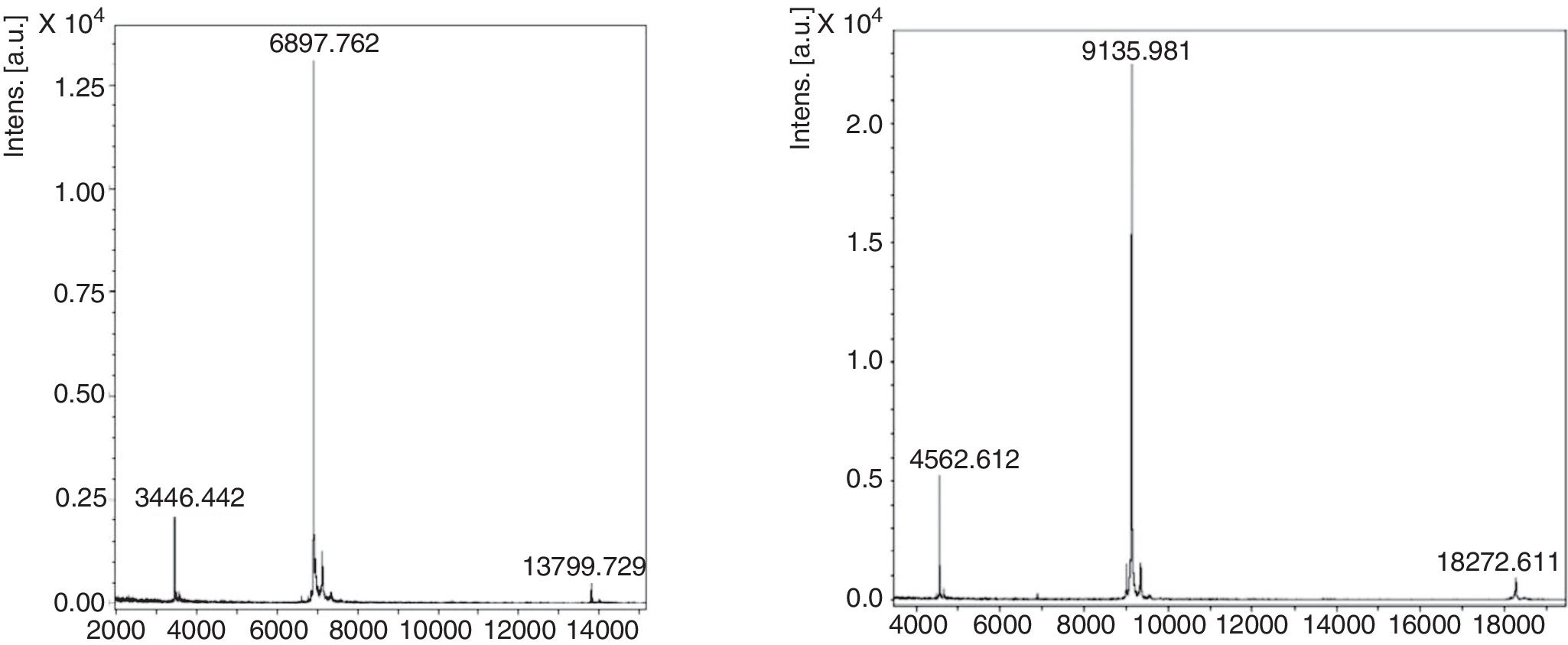
Protein concentration was determined using a DC Protein Assay kit (Bio-Rad, Bradford, UK). The N-terminal amino acid sequences of Pru p 7 and Pru p 3 were determined using a 476A gas-phase protein sequencer (Applied Biosystems, CA, USA). The molecular mass and purity of Pru p 7 and Pru p 3 were also checked using an Autoflex III matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS; Bruker Daltonics, MA, USA). Amino acid sequences were analyzed using the Phytozome and BLAST programs to identify the proteins in the databases.
Skin-prick testA skin-prick test using a bifurcated needle with fresh peach was performed with the prick-to-prick technique. A wheal diameter of ≥3mm was defined as positive.29 The entire peach, including the skin, was used for pricking. Peaches used for the prick test were prepared at each facility.
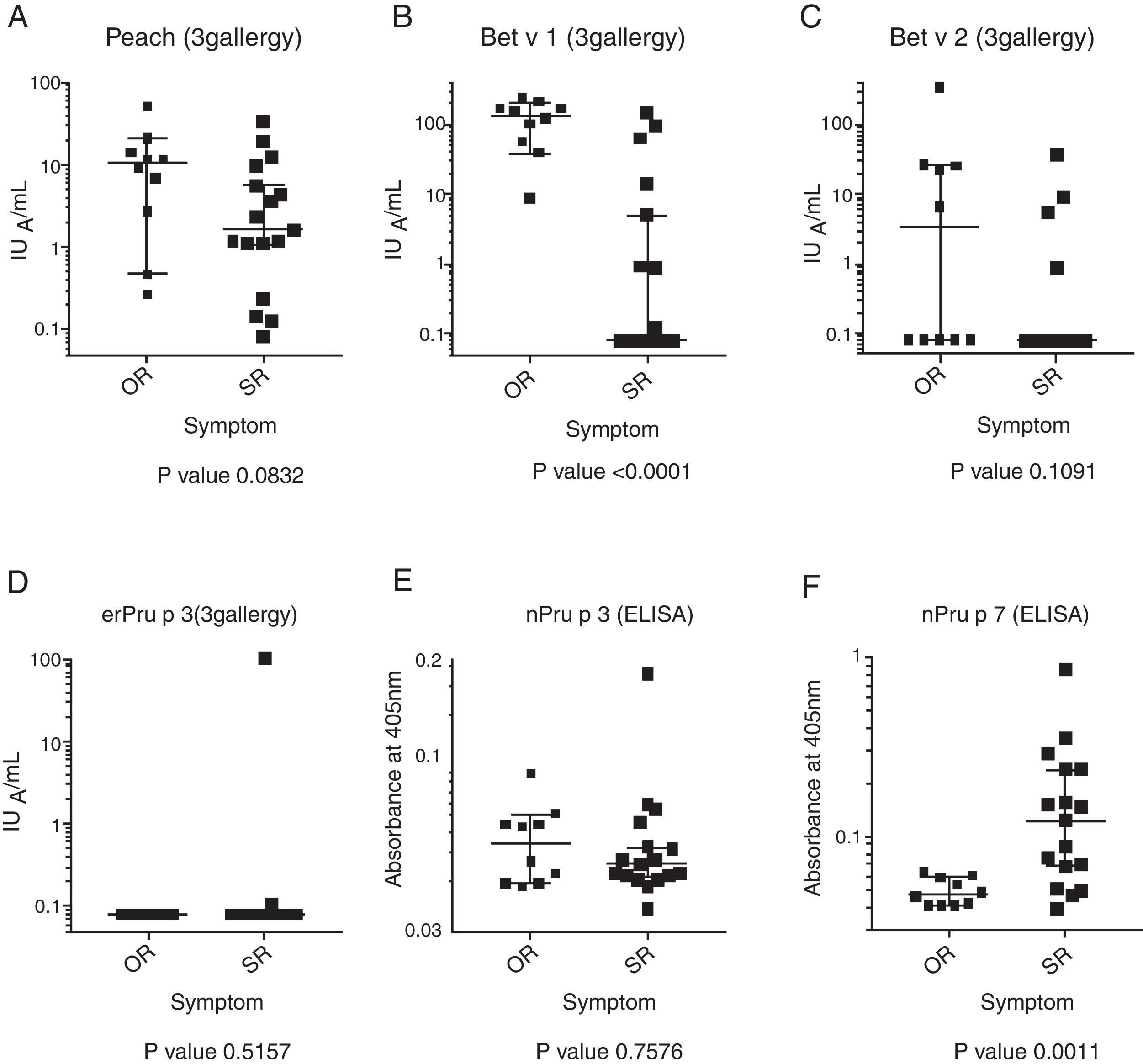
Determination of specific IgE by 3gAllergyThe specific IgE to peach, Pru p 3, Bet v 1, and Bet v 2, were measured by 3gAllergy™ (Siemens Healthcare Diagnostic Inc., Tarrytown, NY, USA). In this study, to simplify calculations, 0.08IUA/mL was substituted for all values <0.1IUA/mL.
Determination of specific IgE in patients’ sera by solid-phase ELISAIgE in patients’ sera specific to Pru p 7 or Pru p 3 was determined using ELISA as described previously with some modifications.6 The wells of a microtiter plate (Nunc A/S, Roskilde, Denmark) were coated with Pru p 7 or Pru p 3 (2μg/mL in PBS) at 50μL/well. After blocking, the individual sera (1:10 dilution with the solution containing 0.1% bovine serum albumin and 0.1% Tween 20 in 10mM Tris–HCl buffer, pH 7.4) were added at 50μL/well and incubated for 1h at 37°C. The IgE Abs bound to the antigen were detected using alkaline phosphatase-labeled anti-human IgE (American Qualex, San Clemente, CA, USA) and p-nitrophenylphosphate (1mg/mL) as the substrate. Absorbance was read at 405nm using a microplate reader (BioRad). Mean+10 standard deviations of the negative control values were defined as positive.
Statistical methodsCorrelations between the clinical symptoms of peach-allergic patients and sensitization to peach, Bet v 1, Bet v 2, nPru p 3, and Pru p 7 were analyzed using the Mann–Whitney U-test. Data sets of nPru p 7, erPru p 7, and prPru p 7 were analyzed for normal distribution using the Kolmogorov–Smirnov normality test. Because most of the data sets were not normally distributed, the non-parametric Friedman test with Dunn's multiple comparison post-test was used. All data were analyzed using GraphPad Prism version 6.0. (GraphPad Software Inc., San Diego, CA, USA).
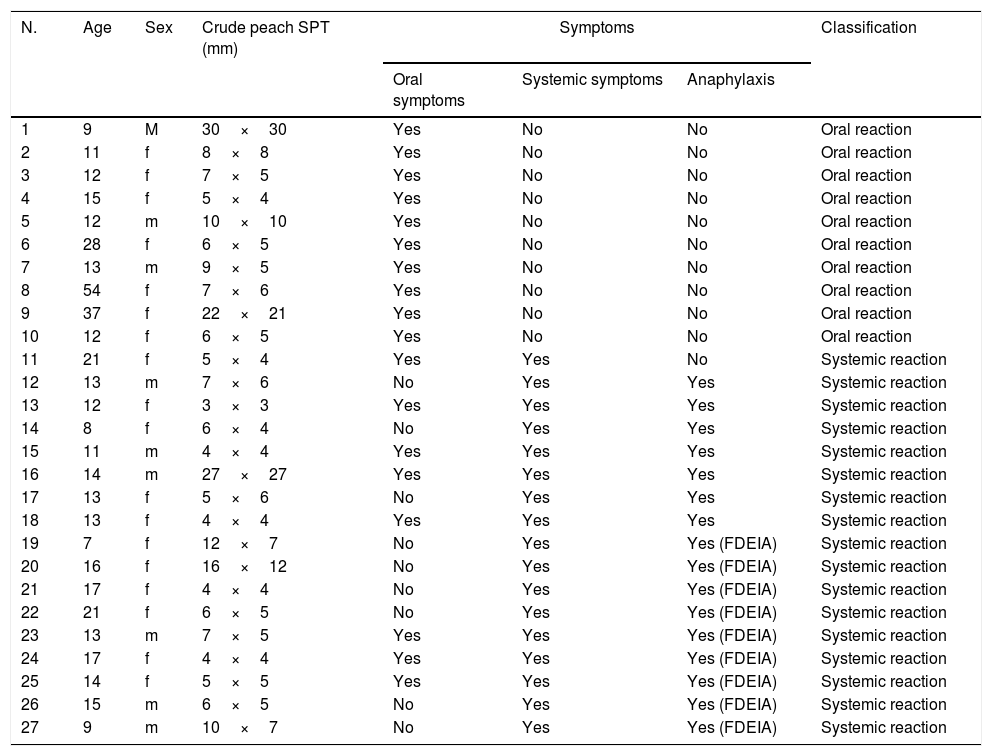
ResultsParticipantsWe examined 27 participants who met the criteria (9 men and 18 women; age range, 7–54 years; median age, 13 years). The participants were classified into the SR (n=17, 6 men and 11 women; age range, 7–21 years; median age, 12.5 years) and OR groups (n=10, 3 men and 7 women; age range, 9–54 years; median age, 13 years) (Table 1).
Background of subjects.
| N. | Age | Sex | Crude peach SPT (mm) | Symptoms | Classification | ||
|---|---|---|---|---|---|---|---|
| Oral symptoms | Systemic symptoms | Anaphylaxis | |||||
| 1 | 9 | M | 30×30 | Yes | No | No | Oral reaction |
| 2 | 11 | f | 8×8 | Yes | No | No | Oral reaction |
| 3 | 12 | f | 7×5 | Yes | No | No | Oral reaction |
| 4 | 15 | f | 5×4 | Yes | No | No | Oral reaction |
| 5 | 12 | m | 10×10 | Yes | No | No | Oral reaction |
| 6 | 28 | f | 6×5 | Yes | No | No | Oral reaction |
| 7 | 13 | m | 9×5 | Yes | No | No | Oral reaction |
| 8 | 54 | f | 7×6 | Yes | No | No | Oral reaction |
| 9 | 37 | f | 22×21 | Yes | No | No | Oral reaction |
| 10 | 12 | f | 6×5 | Yes | No | No | Oral reaction |
| 11 | 21 | f | 5×4 | Yes | Yes | No | Systemic reaction |
| 12 | 13 | m | 7×6 | No | Yes | Yes | Systemic reaction |
| 13 | 12 | f | 3×3 | Yes | Yes | Yes | Systemic reaction |
| 14 | 8 | f | 6×4 | No | Yes | Yes | Systemic reaction |
| 15 | 11 | m | 4×4 | Yes | Yes | Yes | Systemic reaction |
| 16 | 14 | m | 27×27 | Yes | Yes | Yes | Systemic reaction |
| 17 | 13 | f | 5×6 | No | Yes | Yes | Systemic reaction |
| 18 | 13 | f | 4×4 | Yes | Yes | Yes | Systemic reaction |
| 19 | 7 | f | 12×7 | No | Yes | Yes (FDEIA) | Systemic reaction |
| 20 | 16 | f | 16×12 | No | Yes | Yes (FDEIA) | Systemic reaction |
| 21 | 17 | f | 4×4 | No | Yes | Yes (FDEIA) | Systemic reaction |
| 22 | 21 | f | 6×5 | No | Yes | Yes (FDEIA) | Systemic reaction |
| 23 | 13 | m | 7×5 | Yes | Yes | Yes (FDEIA) | Systemic reaction |
| 24 | 17 | f | 4×4 | Yes | Yes | Yes (FDEIA) | Systemic reaction |
| 25 | 14 | f | 5×5 | Yes | Yes | Yes (FDEIA) | Systemic reaction |
| 26 | 15 | m | 6×5 | No | Yes | Yes (FDEIA) | Systemic reaction |
| 27 | 9 | m | 10×7 | No | Yes | Yes (FDEIA) | Systemic reaction |
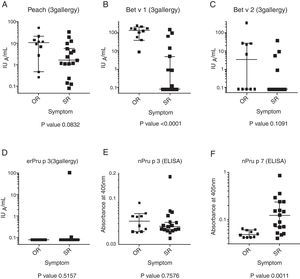
All the participants were positive for the skin-prick test against peach. The level of the specific IgE to Bet v 1 in the OR group was significantly higher than that in the SR group (P<0.0001). The Mann–Whitney U-test showed no significant difference in the level of the specific IgE to peach, Bet v 2, and nPru p 3 between the SR and OR groups (Fig. 1). It was noted that only patient no. 18 in the SR group showed a positive response against nPru p 3, consistent with the results of the 3gAllergy™ test. As reported previously,6 the level of the specific IgE to nPru p 7 in the SR group was significantly higher than that in the OR group (P=0.0011).
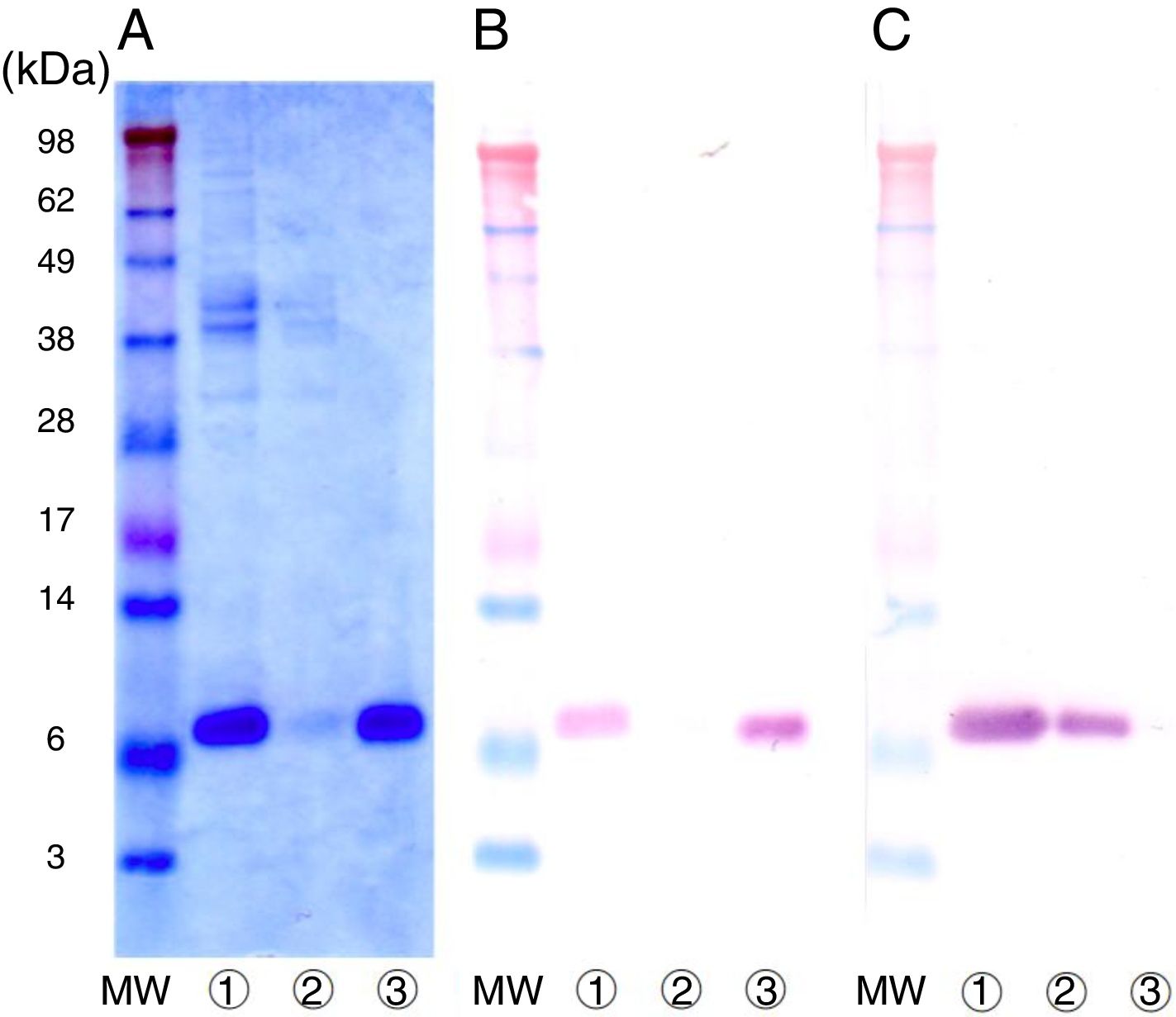
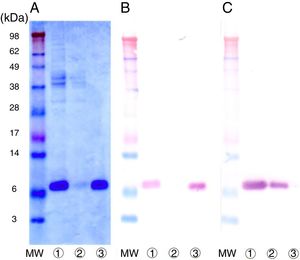
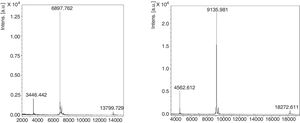
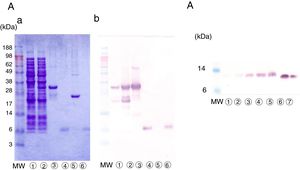
Purification of nPru p 7The crude extract solution was prepared using the cation-exchange resin from fully ripened peaches (Fig. 2, lane 1), and a one-step purification of nPru p 7 was performed using an anti-Pru p 7 mAb column (Fig. 2, lane 3). Pru p 3 was passed through the antibody column (Fig. 2, lane 2). On the basis of the results of western blotting, it was evident that nPru p 7 was completely purified and free from Pru p 3. nPru p 3 was also purified using the anti-Pru p 3 mAb column (data not shown). MALDI-TOF MS analysis also indicated the purity and molecular weight as 6898Da and 9136Da for nPru p 7 and nPru p 3, respectively (Fig. 3).
Purification of nPru p 7 by using the anti-Pru p 7 monoclonal antibody column.
Peach crude extract was applied to the anti-Pru p 7 column. The flow through and the eluate fractions were examined on SDS-PAGE under reducing conditions.
(A) Stained with Coomassie Brilliant Blue, (B) Western analysis by using anti-Pru p 7 monoclonal antibody, (C) Western analysis by using anti-Pru p 3 monoclonal antibody.
Lane
apply (peach crude extract), Lane antibody column flow through, Lane antibody column eluate. MW indicates molecular weight marker (in kilodaltons).We extracted RNA from peaches (P. persica, cultivar Asama–Hakutou strain) before they fully ripened and cloned the cDNA of Pru p 7 using PCR. Isolation and sequencing of the cDNA revealed a 189-bp open reading frame coding for a 63-amino acid polypeptide. The nucleotide sequence of the open reading frame was entirely consistent with that obtained from a database. Moreover, the expected molecular weight was consistent with the value mentioned above obtained by MALDI-TOF MS.
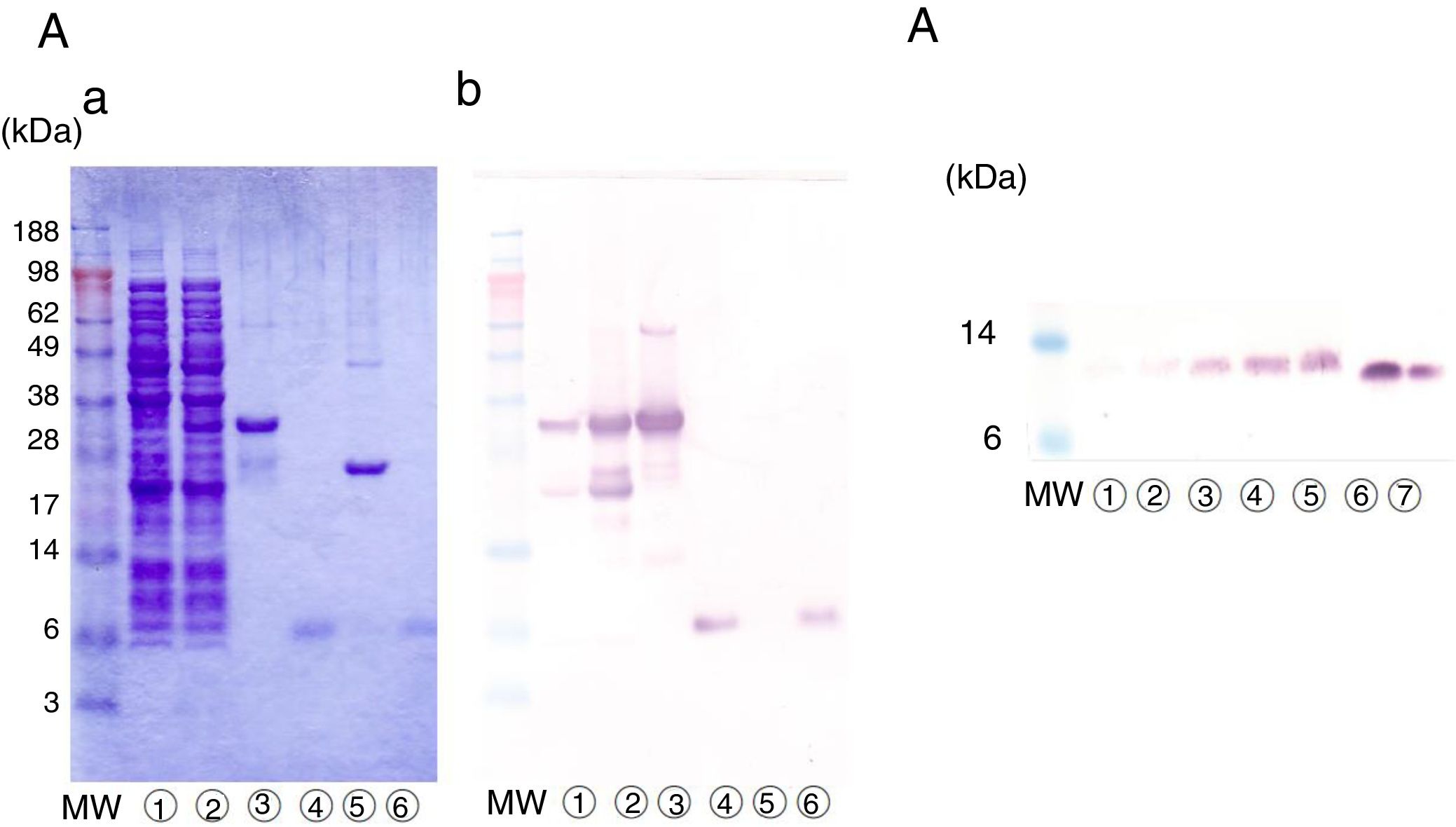
The cDNA clone was first expressed in E. coli as a fusion protein with glutathione S-transferase, with a molecular weight of approximately 34kDa (Fig. 4A-a, lanes 2 and 3). The lysate of the recombinant E. coli was applied to glutathione-Sepharose beads, and erPru p 7 was released from the beads by protease treatment. erPru p 7 was electrophoresed at almost the same position as nPru p 7 in SDS-PAGE (Fig. 4A-a, lanes 4 and 6) and stained with monoclonal anti-Pru p 7 antibody on western blot analysis (Fig. 4A-b). The N-terminal amino acid sequence analysis of erPru p 7 revealed the sequence (GPL)GSSFCDSKCGVRCSKAG, where the original sequence of nPru p 7 (GSSFCD) was correctly expressed following the first tripeptide (GPL) derived from the vector and added by recombination.
Expression of rPru p 7 by E. Coli and P. pastoris.
(A) Detection of erPru p 7 expressed in E. coli.
(a) SDS-PAGE under reducing conditions stained with Coomassie Brilliant Blue.
(b) Western analysis by using anti-Pru p 7 monoclonal antibody.
before induction of expression, after induction of expression, glutathione beads before cleavage, eluate after cleavage, beads after cleavage, nPru p 7.(B) Detection of prPru p 7 secreted in the medium of P. pastoris on western analysis by using anti-Pru p 7 monoclonal antibody.
culture supernatant after 24h, after 48h, after 72h, after 96h, after 120h, nPru p 7 100ng, nPru p 7 50ng.MW indicates molecular weight marker (in kilodaltons).
Pru p 7 cDNA was also expressed in P. pastoris, and prPru p 7 was secreted into the medium by the signal sequence of the vector. Secreted prPru p 7 in the medium was too faint to be detected by Coomassie staining but was visible on western blot analysis (Fig. 4B). Secreted prPru p 7 migrated slightly slower than nPru p 7 probably because of the addition of the extra pentapeptide (EAEAY) at recombination. The N-terminal amino acid sequence analysis of prPru p 7 revealed the sequence (EAEAY) GSSFCD, where the original sequence of nPru p 7 was correctly expressed following the first pentapeptide.
To compare the reactivities of erPru p 7 and prPry p 7 with serum IgE in peach-allergic patients, erPru p 7 and prPry p 7 were purified with mAb columns, similar to nPru p 7. As the reference, Pru p 3 was cloned, and erPru p 3 was expressed in E. coli and purified using an anti-Pru p 3 mAb column.
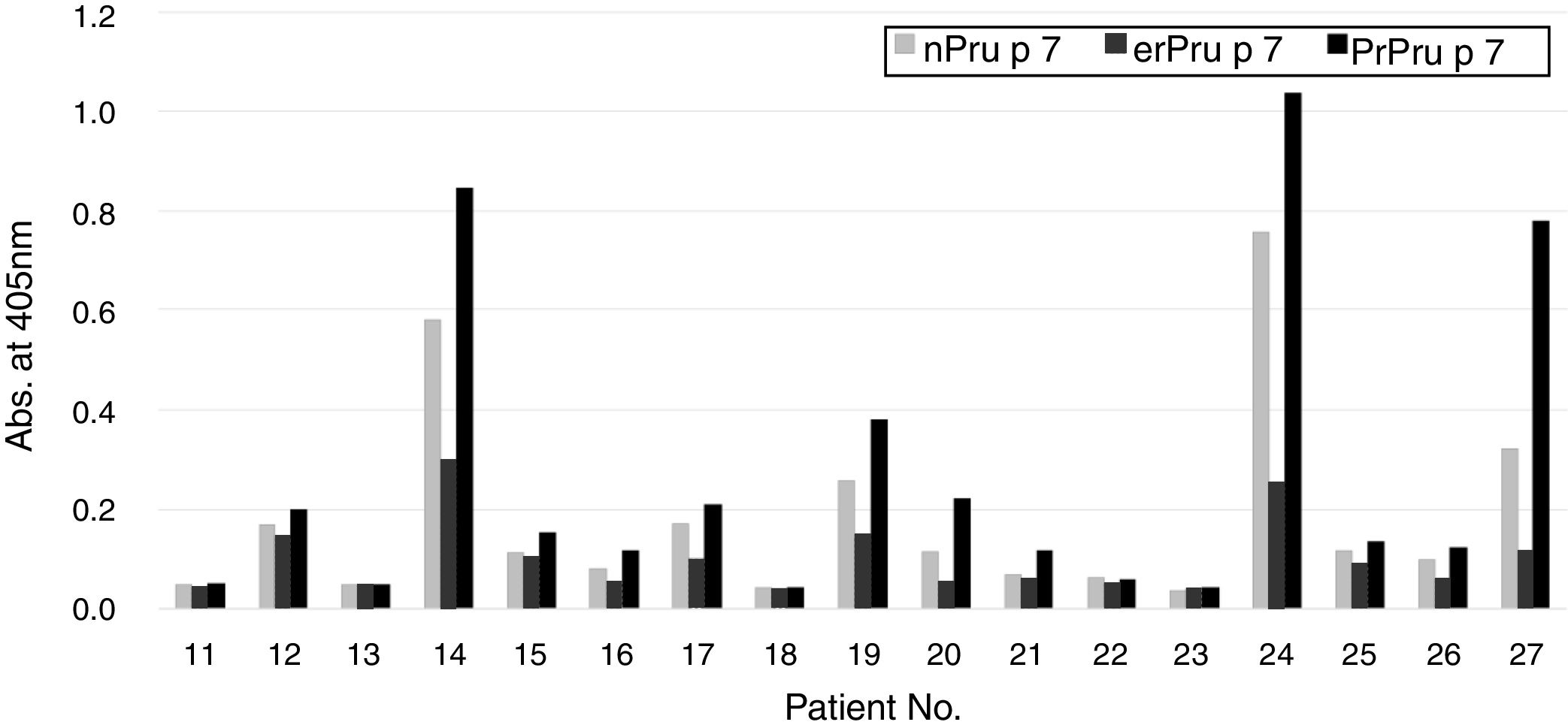
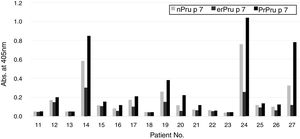
Reactivity of patients’ IgE to nPru p 7and rPru p 7 in ELISAFinally, in the SR group, we compared the reactivity of patients’ IgE to nPru p 7 and rPru p 7 using ELISA (Fig. 1). Because the data did not exhibit normal distribution by the Kolmogorov–Smirnov test, we decided to perform non-parametric analysis. First, a significant difference was observed between each antigen by the Friedman test (P<0.05). Then, Dunn's multiple comparison post-test revealed that a significant difference was not observed between nPru p 7 and prPru p 7 (P>0.05) but was observed among nPru p 7 and erPru p 7 (P<0.05), erPru p 7 and prPru p 7 (P<0.001) (Fig. 5).
Reactivity of patients’ IgE to natural and recombinant Pru p 7 in ELISA.
IgE in the sera of the systemic group (No. 11–27 in Table 1) was determined by ELISA with nPru p 7, erPru p 7, prPru p 7.
Pru p 7 is an allergen found in peach and has recently been reported as a marker related to SRs to this fruit.6,15 To use Pru p 7 as the target of CRD, its qualitatively, quantitatively, and economically stable supply is necessary. Therefore, we assessed rPru p 7 for potential use in the worldwide standardization of the diagnosis for peach allergy.
Pru p 7 is a 7-kDa cysteine-rich protein with 12 cysteines of 63 amino acids, which corresponds to 19% of the total residues.15 The rigid structure formed by the 6 disulfide bonds endows Pru p 7 with heat stability, protease resistance, and allergenicity. There are respective mAbs that recognize the reduced or non-reduced form of Pru p 7. Therefore, the disulfide bonds appear to be extremely important for the structure of expressed rPru p 7.17 Because Pru p 7 was originally a secreted protein, it was expected that the secretion of rPru p 7 into the culture medium through the yeast endoplasmic reticulum system would help to make its structure closer to that of nPru p 7 more than its expression in the inclusion bodies of enteric bacteria would.26
As expected above, statistically different reactivity of serum IgE was not shown between nPru p 7 and prPru p 7. The differences between the sequences of rPru p 7 and nPru p 7 are the presence of additional N-terminal tri- and penta-peptides in erPru p 7 and prPru p 7, respectively. Because the N-terminal of Pru p 7 sticks out of its core,17 the N-terminal differences do not seem to severely affect the 3D structure of Pru p 7. Because Pru p 7 has no glycosylation site in yeast and because of its molecular weight (Fig. 4), prPru p 7 appeared to be expressed as a simple protein. Therefore, the differences in this reactivity are thought to be attributable to the nature of the SS bonds. Taking these observations into account, it was appropriate to select the P. pastoris system not only for Pru p 7 but also for general recombinant proteins.20,22–26 In addition, the P. pastoris system enabled us to obtain high-purity products in the medium and stable transgenes by homologous recombination.22
From the clinical standpoint, we reconfirmed earlier findings that over half of the systemic patients for peach allergy are positive for Pru p 7 by ELISA in Japan.6 Regardless of the use of native or recombinant materials, this response should also be estimated using other in vitro tests, such as histamine release and basophil activation. Furthermore, studies using large numbers of patients with various symptoms in Japan and in other countries are necessary to clarify the relevance of gibberellin-regulated protein and LTP to fruit and vegetable allergies.
Although Pru p 3 has been reported as the major component for systemic peach allergic patient in Europe,13,30,31 patients sensitized to Pru p 3 are very rare in Japan.6,16 In this study, IgE from only one systemic patient (no. 18 in Table 1, Fig. 1D) definitely reacted to nPru p 3 and rPru p 3, which indicated that LTP allergy is rare but present in Japan. This difference may be caused by eating habits; the Japanese usually peel a peach before eating it, whereas Europeans eat the entire peach.14 Our preliminary experiments using sandwich ELISA and mAbs showed that 0.6 and 41μg/g of Pru p 3 were present in pulp and skin and 23 and 3.4μg/g of Pru p 7 were present in pulp and skin, respectively. Although somewhat dependent on cultivars and markedly on the maturity of the peach, Pru p 3 is localized in skin and Pru p 7 in pulp.17 The establishment of a proper assay system for Pru p 7 is expected to contribute to the prevention of peach allergy through quality control and allergen labeling.
Finally, in this study, we succeeded in cloning Pru p 7. Various applications, such as epitope and structural analysis, can be expected using gene manipulation. Several improvements, including the production of hypo-allergenic and anti-pathogenic variants, such as transgenic potato, are also expected.32
In conclusion, this study revealed that rPru p 7 expressed by P. pastoris exhibits reactivity in ELISA comparable with that of nPru p 7.
Trial registrationNot applicable.
Funding sourceNone.
Conflict of interestNone.
We are grateful to the doctors of the Fruits Allergy Component Study Group for patient recruitment, Teshigawara from the Department of Pediatrics, Fujita Health University, for her technical support in the experiments, and Siemens for performing the measurement of IMMULITE® 2000 3gAllergy™ for allergen components.