INTRODUCTION
Atopy results from the interaction between genetic and environmental factors. It is associated with rised serum IgE levels. IgE are released from B cells, enhancing the degranulation of mast cells with FcεRI receptors blocked by IgE/allergen complexes. Released histamine provokes the cytokines production including IL-4, which activates pre Th helper cells and their transformation to Th2 cells1. Overexpression of the IL-4 gene may affect Th2 differentiation and class switching to IgE. Several authors have linked the IL-4 gene located on chromosome 5q to atopy and bronchial hyperresponsiveness2 Marsh et al in USA, Meyers et al and Postma et al in Dutch families described genetic loci in chromosome 5q suspected to be linked with total serum IgE levels3-5. The (-590 C/T) Il-4 gene promoter polymorphism was reported to be associated with elevated levels of total serum IgE and with asthma symptoms6 The gene for βsubunit of FcεRIβ receptor was also indicated as a candidate gene for atopy7,8 It was shown that variants of FcεRIβ gene, coding variant Ile181Leu and non-coding INT2, are associated with an increased risk of atopy and asthma9,10. Some authors underlined FcεRIβ gene linkage to atopy by its high prevalence in atopics and maternal inheritance11,12. In our study we examined the genetical, immunological and clinical features of atopy in a case-control studies in a Polish population sample, investigating (590 C/T) Il-4 gene promoter polymorphism, polymorphism of FcεRIβint2 and mutation Glu237Gly in FcεRIβ gene.
SUBJECTS
All case and control subjects were the residents of the urban area of Lodz in Poland and they were aged 18-45. Case group consisted of 98 unrelated atopic patients with clinical manifestations of atopy like asthma, allergic rhinitis or asthma and allergic rhinitis. Patients enroled in the study came from the Medical University Hospital outpatient clinic where they had been treated at last for one year. Also 87 non-atopic, non-asthmatic, unrelated control subjects, citizens of Lodz, were included in the study.
MATERIALS AND METHODS
All of case and control subjects underwent the following procedures: Severity of airway dysfunction was established using spirometric measurements. Spirometry (FEV1 > 20 % in asthmatics) was done in all individuals enrolled in the study. Atopy status was defined as described in other papers: at least one skin prick test (spt) had to be positive to a common allergen, a positive RAST test to one or more allergens, elevated circulating total IgE and confirmed clinical symptoms of allergy13,14. To determine atopic status common spts to hdm (Dermatophagoides pteronyssimus) and mgp (Artemisia, Timothy grass, Rye grass) were done and a positive spt was defined as 3 mm greater than a negative control. Case subjects were enrolled in the study only under all above restrictions. We excluded patients with negative spts and elevation of the total IgE in that way.
10 ml of whole blood sample was taken with a closed EDTA coated system. Total serum IgE was measured by Pharmacia CAP system. Specific IgE responses to hdm and mgp were determined by radioallergosorbent test (RAST). Total IgE levels greater than 200 IU/mL and RAST classes greater than 2 were considered as positive tests. Blood samples were transported to Lung Research Laboratory, Osler Chest Unit, Churchill Hospital, Oxford, UK. Genomic DNA was isolated from peripheral white blood cells, using a commercial kit (Iso Quick, Microprobe Corporation, Garden Grove, USA).
PCR reaction for FcεRIβint2 was performed in a total volume sample of 30μl. PCR mixture consisted of 20 μM of 3' and 5' primers: gAATggCCAACAggAg TgAAggAT and CAAgTACAgAgCAgACAACTg, 200ng of template DNA, 25nM of each dNTPs, PCR buffer, 2U Taq polymerase (Gibco BRL) and 100μl of mineral oil. PCR reaction consisted of: preliminary cycle: 94 °C for 5 min, 60 °C for 1 min and 72 °C for 2 min and 34 cycles: 94 °C for 30 sec, 60 °C for 30 sec, 72 °C for 1 min.
PCR reaction for the (590 C/T) Il-4 gene promoter polymorphism was performed in a total volume of 15μl. PCR mixture consisted of 20μM of 3' and 5' primers: ACTAggCCTCACCTgATACg and gTTgTAATg CAgTCCTCCTg, 100 ng of DNA template, 25 nM of each of dNTPs, PCR buffer, 2.5 U Taq polymerase and 100 μl of mineral oil. Amplification performed in a thermal cycler was preliminary cycle: 94 °C for 5 min and next 32 cycles: 94 °C for 30 sec, 60 °C for 30 sec, 72 °C for 5 min.
The mutation Glu237Gly was investigated using an allele refractory mutation screening (ARMS) test. The reaction mixture consisted of: 200ng of DNA template, 0.5μM. of primers:
1a:TggCCAgCTAgTCTggTTTggTTTTCTggA
1b:ggAgCATATTAAggTggACAgAAgCAgCAg
2a:ATTCAgCTACTTACAgTgAgTTggAAgAC CCAggCgg
2b:CACgTgATTCTTATAAATCAATgggAggAgA CAATT
and 200μM. each of dNTPs, 2U of Taq polymerase and PCR buffer up to total volume 50μl. ARMS reaction consisted of: hot start" PCR, pre-cycle at 95 °C for 5 min and next 35 cycles:
94 °C for 1 min, 60 °C the (590 C/T) Il-4 gene promoter polymorphism for 2 min, 72 °C for 2 min, last cycle ended with elongation at 72 °C for 10 min.
Amplification products (5μl each) were digested with restriction enzymes: 1U of Rsa 1 for the FcεRIβint2 and 2U of BsmF1 for the (590 C/T) Il-4 gene promoter polymorphism. Restrictions were performed for 2 hrs at 37 °C and stopped by the agarose gel loading buffer (5 μl). The digested products and DNA molecular weight marker were loaded onto agarose gel (a 2 % (w/v) agarose/1xTBE gel), containing of 50ng/ml ethidium bromide. In the case of ARMS-PCR, amplified products were loaded onto a 4 % agarose gel (3:1, Nu Sieve GTG: LMP agarose). Electrophoresis was performed at 70V for 90 min. DNA fragments were visualised in UV light and photographed.
RESULTS
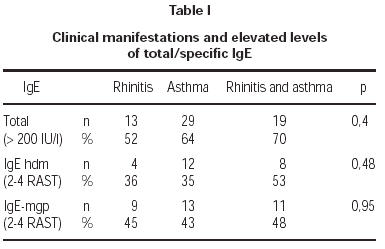
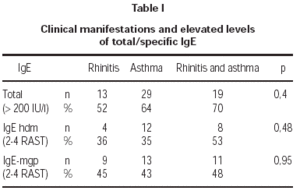
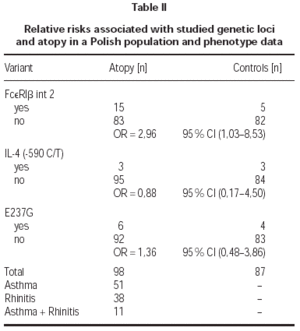
Total serum levels were rised (> 200 IU/ml) in 62 % of atopics and in 9 % of controls. Atopy was strongly linked to elevated total IgE concentrations in patients (OR = 16,28). A medium level of total IgE in atopy group was 240 IU/ml, in non-atopics: 26,6 IU/ml. There was no linkage between the high total and specific IgE levels and clinical manifestations of atopy like atopic asthma or allergic rhinitis (table I).
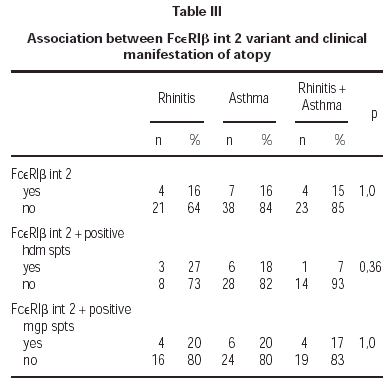
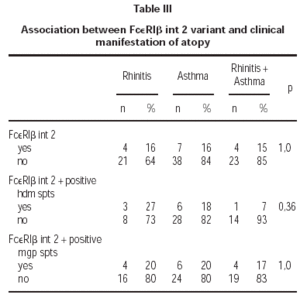
The (590 C/T) Il-4 gene promoter polymorphism were not associated with atopy in this Polish population sample (OR = 0,88; table II). Also there was no correlation of mutation Glu237Gly in exon 7 of FcεRIβ with atopy (OR = 1,36; table II). Additionally we found no homozygous individuals for that variant in Polish population. We found statistically significant correlation between FcεRIβint2 and atopy (OR = 2,96; table II). That variant was not significantly associated with symptoms of atopic asthma or allergic rhinitis (table III).
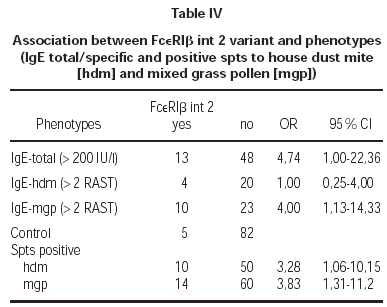
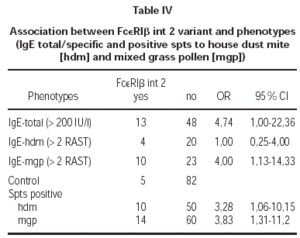
There was a correlation between elevated total IgE levels and the prevalence of FcεRIβint2 variant (OR = 4,74; table IV). We found significant correlation between positive spts to hdm (OR = 3,28; table IV) and mgp (OR = 3,83; table IV) and the FcεRIβint2 variant.
Additionally we observed a high association between the presence of polymorphic FcεRIβint2 variant in atopics and isolated positive spts to mgp (OR = 2,48) and positive spts to mgp and hdm (OR = 5,47). The elevated specific IgE for mgp and positive spts to mgp were correlated with that variant (OR = 4,0; table IV).
We did not find association of FcεRIβint2 with positive spts to hdm and rised IgE for hdm (OR = 1,0; tab.). Also there was no correlation between studied variant FcεRIβint2, positive spts to hdm or mgp and clinical manifestations of atopy (p = 1).
DISCUSSION
In early studies of the (590 C/T) Il-4 gene promoter polymorphism, Rosenwasser at al postulated its influence on overproduction of total IgE and overexpression of transcriptional factors in lymphocytes T resulting in a higher IL-4 concentration6,15 Blumenthal et al indicated uncertain role of genetic loci in the candidate region 5q 31-33 with IL-4 gene and genetic regulation of atopy. They underlined the importance of randomised control groups for genetic studies16. Data published by Walley and Cookson also did not show statistically significant association of the (590 C/T) Il-4 gene promoter polymorphism and asthma or atopy in an Oxfordshire sample and a weak correlation with hdm specific IgE and wheezing in the Busselton population, Australia17. There was no association of that marker and total IgE concentration. Noguchi et al suggested that the (590 C/T) Il-4 gene promoter polymorphism may be associated with the development of asthma in Japanese children but it did not affect total IgE levels18 [In recent studies, Australian investigators indicated the haplotype (590C/-34C) of Il-4 gene promoter polymorphism is strongly linked with predisposition to early-onset of atopic eczema19. Wenner et al showed that IL-4 gene activity may be controlled by additional elements outside of IL-4 gene promoter and that more precise identification of the allelic forms is necessary for further genetic investigation20. We did not confirm linkage between that variant and atopy (OR = 0,88), also we did not find any correlation with the level of total or specific IgE in a Polish sample. Our results are confirmed by data presented by Hijazi and Haider who did not find association between asthma and the (590 C/T) Il-4 gene promoter polymorphism in Kuwaiti Arabs. They published interesting study of coding variant of the FcεRIβ gene and did find an association of the homozygous variant Leu181/Leu183 with severity of asthma in the same population21,22 Therefore investigations of that coding variant in a Polish population would be of worth.
Hill et al concluded that FcεRIβLeu181/Leu183 variant may be inherited maternally and a genetic risk factor for atopy and bronchial hyperresponsiveness. A strong correlation between positive spts to hdm and mgp, elevated specific IgE levels and bronchial hyperresponsiveness tests in children with asthma or rhinitis was established23. Other authors also described that variant as maternally inherited24,25.
The coding variant Glu237Gly in the FcεRIβgene has also been studied in various populations with asthma and atopy. The results were controversial for association of that variant with atopy or its clinical manifestations. Its association with atopy and bronchial hyperresponsiveness was found in an Australian population and was published by Hill et al26. The variant Glu237Gly was associated with atopic asthma, particularly with childhood asthma and very high total serum levels in Japanese27. Recently, Laprise et al detected a strong correlation between atopy and Glu237Gly in French-Canadian population28. Weak evidence for linkage to that variant was presented by Simon et al in British atopic asthmatics29. Nagata et al showed that Glu237Gly is involved in the development of nasal allergy. That mutation was associated with rised total IgE levels and specific IgE for hdm and Japanese cedar pollen30. In our study we did not find homozygous individuals as Japanese did. There was no correlation with atopy and its phenotypes in a Polish group.
We also investigated FcεRIβint2 variant and we found a strong association with atopy and this is the first documentation of this in an Eastern European population. Intron 2 polymorphism was closely correlated with elevated total IgE levels and positive spts to hdm and mgp. The study of patients with the present polymorphic variant showed linkage with positive spts to mgp and elevated specific IgE for mgp and no linkage with positive spts to hdm and IgE for hdm. Similar data were obtained by Shirakawa et al. They described correlation between locus INT2 and high total IgE and specific IgE levels for mgp31. They also indicated the influence of investigated loci on the development of atopic asthma, rhinitis and eczema but used very high total IgE levels (> 400 kU/mL) for randomisation. We did not find a correlation of variant FcεRIβint2 with clinical features of atopy. A recent study of Spanish group showed that variant FcεRIβint2 was associated with rised specific IgE for olive tree pollen (ole e 1)32. The associations indicate that the FcεRIβgene is a significant locus for atopy. Variants are likely to disturb its function but further studies are needed to clarify the mechanism of enhanced IgE sensitisation to common allergens.