The pathogenesis of chronic spontaneous urticaria involves interplay between the genetic and environmental factors, most of which is still poorly understood. It is well-recognized that 30–40% of chronic spontaneous urticaria is autoimmune in nature. Chronic autoimmune urticaria is caused by anti-Fc¿R1β and less frequently, by anti-IgE auto antibodies that lead to mast cell and basophil activation, thereby giving rise to the release of histamine and other proinflammatory mediators. We investigated the association between SNP loci in Fc¿R1β and chronic spontaneous urticaria and to see its relation with serum IgE levels in Kashmiri population.
MethodsThe autologous serum skin test was used as a screening test for chronic autoimmune urticaria. PCR-RFLP was used to detect the genotype of the SNP loci. Serum IgE levels were assessed by ELISA kit.
ResultsNo significant difference was found between the study population and control group in genotype distribution (wild and variant) among Fc¿R1β loci (P value=0.06, odds ratio=0.29). The frequency of Fc¿R1β (C109T) in autologous serum skin test positive chronic autoimmune urticaria patients with the CT genotype was found to be statistically non-significant when compared with the wild genotype (P=0.35). Carriers of Fc¿R1β (T allele) had a more significant risk of developing CAU than those with C allele (P=0.01). In our population serum total IgE levels did not find any statistical significance with regard to ASST positive & ASST negative patients (P=0.26).
ConclusionsThere is statistically no significant association between Fc¿R1β gene polymorphism and CSU in Kashmiri population; however, there is a probability of developing CSU in patients carrying Fc¿R1β T allele. Furthermore, serum total IgE levels had no significant association with the development of CAU.
Urticaria involves the superficial portion of the dermis, presenting as well-circumscribed wheal with erythematous raised serpeginous borders and blanched centers that may coalesce to become giant wheals. Chronic urticaria is one of most common chronic inflammatory disorder prevalent in our society, defined as urticaria persisting daily or almost daily for more than six weeks. Chronic urticaria causes deterioration of quality of life.1 It includes physical urticaria, cholinergic urticaria, chronic idiopathic urticaria and urticarial vasculitis. Up to 55% of the patients with chronic urticaria have an idiopathic cause. In recent years a significant number of patients (30–40%) with CSU have been reported to have an autoimmune basis for their disease including autoantibodies to IgE (5–10%) or, more commonly, to the (α-chain) of Fc¿R1 (35–45%).2 These autoantibodies have been shown to activate blood basophils and cutaneous mast cells in vitro, with enhancement of basophil activation by complement and release of C5a. Unfortunately antibody binding tests (viz. ELISA) can yield positive tests in many autoimmune diseases, normal subjects, or patients with other forms of urticaria, but most such sera are nonfunctional and cannot be used to diagnose CAU. At this point of time, gold standard for detecting clinically relevant autoantibodies to Fc¿R1 is functional in vitro donor basophil histamine release assay (BHRA) and their specificity is confirmed by immunoassay (Western blot or ELISA), where these tests are available.3
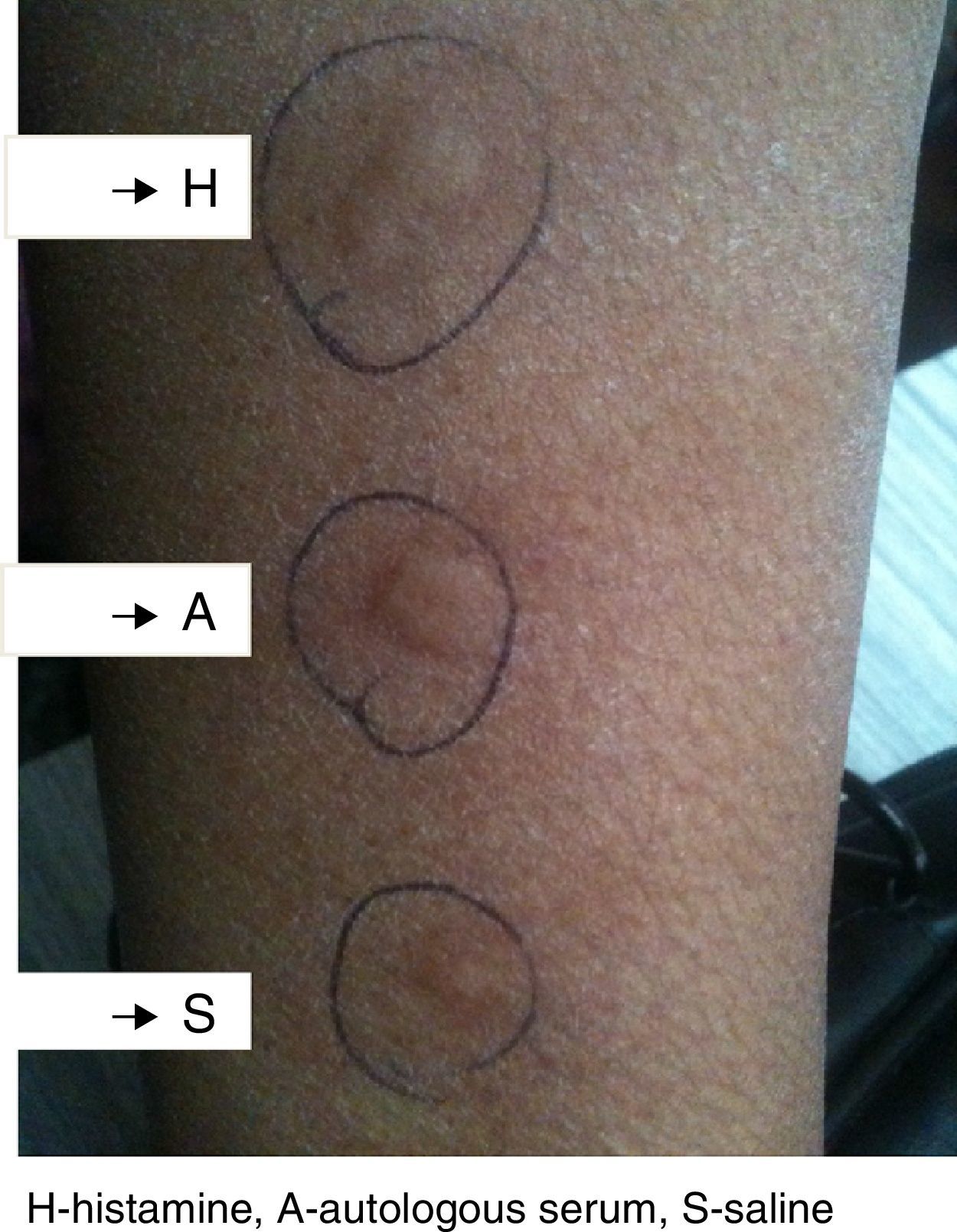
In autologous serum skin test (ASST), autologous serum injected into patients own skin can induce a wheal and flare reaction involving mast cell activation, and removal of the antibody leads to remission.4 CAU has been reported to occur in children as well.5 It is often not possible to distinguish CAU from those without autoantibodies, clinically or histologically. The ASST is used as a screening test. Grattan et al., reported that the histological features of a positive ASST resemble an IgE-mediated late phase reaction.6 The ASST is a useful tool for picking up patients with circulating wheal producing factors in CSU. However, its specificity as a screening test for presence of functional anti-Fc¿R1 is moderate while the sensitivity is high enough to safely exclude autoimmune urticaria and confirmation by demonstration of histamine-releasing activity in the patient's serum may be needed for establishing this diagnosis.
To release histamine from mast cells, the key step is a cross-linking of allergens with IgE, which is bound with the Fc¿R1β located on their surface.7 The receptor is a tetrameric complex composed of an alpha, a beta and two disulphide-linked gamma chains.8 While the genes for the alpha and gamma subunits are both located on human chromosome 1,9 the beta gene is located on 11q13 and spans about 10kb and contains 7 exons.10 Histamine release via autoantibodies against Fc¿R1β was noted in the pathogenic mechanism of CSU.11,12 Several investigators demonstrated a positive association of gene polymorphism of the β chain of Fc¿R1β with high serum total IgE level, atopy or the asthma phenotype.12–15 In this study, we analyzed known single nucleotide polymorphism (SNP) of Fc¿R1β in CSU patients, compared with other ASST positive CAU and in normal healthy controls in Kashmiri population. Furthermore we observed association of ASST positive CAU with serum total IgE levels.
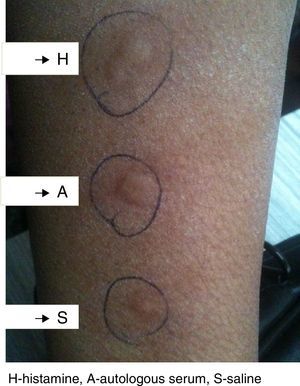
Methods and materialsThe study included 120 patients of CSU and equal number of healthy controls. These patients were enrolled by the allergy clinic of department of Immunology and Molecular Medicine, Sher-i-Kashmir Institute of Medical Sciences (SKIMS), Srinagar, India. All the subjects were native, with normal controls having no history of allergy, atopy, drug hypersensitivity or family history of urticaria. All the subjects were recruited from the general population and informed consent was taken, which was then approved by SKIMS ethical committee. CSU was defined as the daily or near daily presence of pruritic wheal for more than 6 weeks without underlying etiology. Autologous serum skin test (ASST) was performed by injecting autologous serum intradermal to volar aspect of same patient's forearm, and the injected skin was examined for wheal formation 30min later. Positive and negative controls with intradermal histamine and saline injections were carried out on the adjacent skin. The test was considered positive if the serum induced wheal was at least 1.5mm greater than the saline wheal, (Fig. 1). The form of urticaria with positive ASST is characterized as autoreactive. In all the subjects, serum total IgE levels were estimated by performing Enzyme linked immunoassay (ELISA) kit (Fortress diagnostics, UK) following manufacturer's protocol.
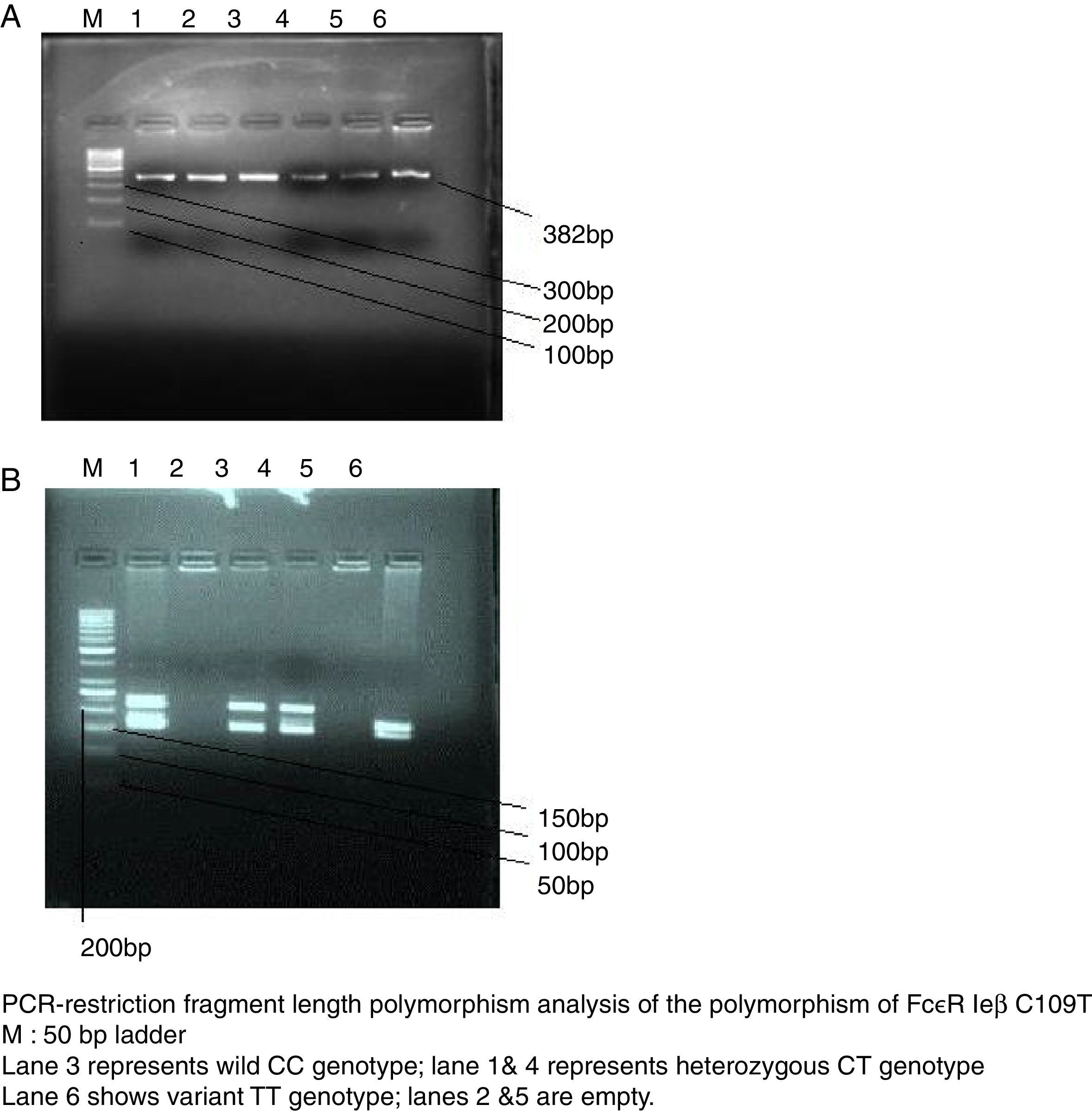
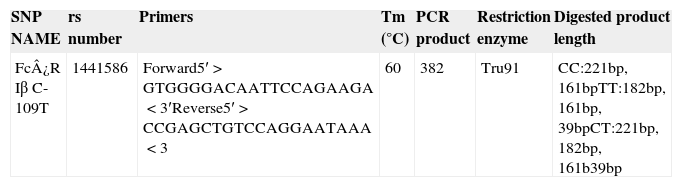
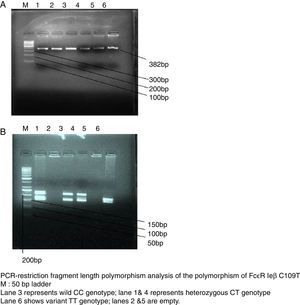
SNP genotyping for allele frequencies of the candidate geneGenomic DNA was isolated from peripheral blood using phenol–chloroform method. Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) was applied to detect the genotype of SNP loci, The reference SNP(rs) number, primers, melting temperature, PCR product length, restriction enzyme, and digested products length of Fc¿R1β SNP loci(C109T) is shown in Table 1. PCR thermal cycler (BioRad) was used for performing reaction with proof reading polymerase by using net volume of 10μL, including 2.05μL of a commercial PCR master mix (Biotools B & M Labs, S.A.), 5pmol of each primer (Sigma–Aldrich Chemicals Pvt. Ltd., India), and 10ng of genomic DNA. The negative control was used to monitor contamination. Cycling conditions included 1 cycle at 95°C for 5min, 40 cycles at 95°C for 30s, annealing temperature for 45s, 72°C for 1min, and a final extension at 72°C for 10min. PCR-amplified products were detected by 2% agarose gel electrophoresis (Fig. 2a). One sample was not included in the study due to the non-amplification of the candidate gene. A restriction enzyme (Tru91) with restriction site TGAA>TTAA was used to digest the PCR products of the loci. A 10-μL reaction mixture containing 5 units of restriction enzyme and 5-μL of PCR products was incubated for 16h. Digested products were run on 2.5% agarose (High EEO, Highmedia Labs, India) in 1× TAE buffer. The electrophoresis was carried out at 80V and 200mA and photographed using the automatic digital gel imaging system (Fig. 2b). CC genotype was considered as homozygous wild (normal), TT as homozygous variant & CT as heterozygous variant.
Overview of the SNP genotyped in our study using PCR-RFLP.
| SNP NAME | rs number | Primers | Tm (°C) | PCR product | Restriction enzyme | Digested product length |
|---|---|---|---|---|---|---|
| Fc¿R Iβ C-109T | 1441586 | Forward5′>GTGGGGACAATTCCAGAAGA<3′Reverse5′>CCGAGCTGTCCAGGAATAAA<3 | 60 | 382 | Tru91 | CC:221bp, 161bpTT:182bp, 161bp, 39bpCT:221bp, 182bp, 161b39bp |
Differences in genotype distribution between the experimental and control group were analyzed using χ2 Test. A P value of 0.05 or less was considered statistically significant. All statistical analyses were performed using SPSS version 13.0 (Chicago, USA).
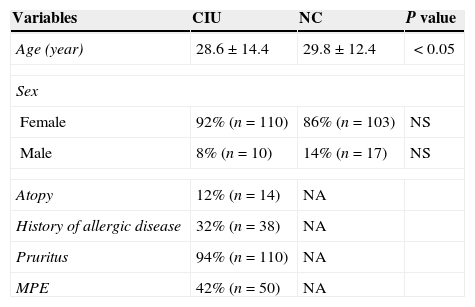
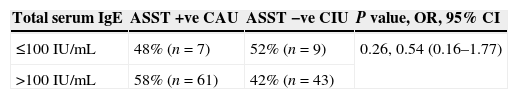
ResultsIn this study 120 CSU patients and equal number of healthy controls were included. The clinical characteristics of study subjects are summarized in Table 2. The age of patients ranged from 7 to 65 years, the mean age was 28 years with females out-numbering males, 11:1 ratio. Atopy was found in 12% of patients, 32% patients revealed the history of allergic disease, 94% patients were having pruritic rashes. As far as the total serum IgE in patients is concerned, the patients were divided into two groups. Those having the total IgE concentration of ≤100IU/mL (group I) and group II having the total IgE concentration of >100IU/mL. In the group I, 48% (n=7) patients were ASST positive and 52% (n=9) patients were ASST negative. In the group II, 58% (n=61) patients were ASST positive and 42% (n=43) patients were ASST negative. Thus, total IgE levels in ASST positive and negative patients varied; there existed no statistical significance in total IgE levels between ASST positive and ASST negative patients among the two groups (P=0.26, OR=0.54, 95% CI=0.16–1.77) as shown in Table 3.
Clinical characteristics of the study subjects.
| Variables | CIU | NC | P value |
|---|---|---|---|
| Age (year) | 28.6±14.4 | 29.8±12.4 | <0.05 |
| Sex | |||
| Female | 92% (n=110) | 86% (n=103) | NS |
| Male | 8% (n=10) | 14% (n=17) | NS |
| Atopy | 12% (n=14) | NA | |
| History of allergic disease | 32% (n=38) | NA | |
| Pruritus | 94% (n=110) | NA | |
| MPE | 42% (n=50) | NA | |
CIU: chronic idiopathic urticaria, NC: normal control, NA: not applicable, NS: non significant, MPE: maculopapular exanthematous rash.
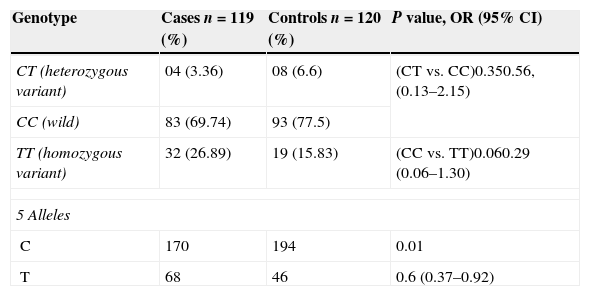
Statistically non-significant differences (P>0.05) were found between the cases and the control group (C/C genotype) in the genotype distribution of Fc¿R1β C109T in loci (Table 4). In our study, we found the frequency of C/C genotype was 69.74% (n=83), T/T genotype was 26.89% (n=32) and that of CT was 3.36% (n=04) in cases, where as it was 77.5% (n=93), 15.83% (n=19) and 6.66% (n=08) in healthy controls, respectively. The frequency of Fc¿R1β C109T, T/T genotype in the ASST +ve CIU group was found statistically non-significant with that of the control group (P=0.06, OR=0.29, 95% CI=0.06–1.30). Similarly, the frequency of Fc¿R1β (C109T) in ASST +ve CAU patients with the CT genotype was found to be statistically non-significant when compared with the wild genotype (P=0.35, OR=0.56, 95% CI=0.13–2.15).
Analysis of association between Fc¿R 1β C-109T and CIU.
| Genotype | Cases n=119 (%) | Controls n=120 (%) | P value, OR (95% CI) |
|---|---|---|---|
| CT (heterozygous variant) | 04 (3.36) | 08 (6.6) | (CT vs. CC)0.350.56, (0.13–2.15) |
| CC (wild) | 83 (69.74) | 93 (77.5) | |
| TT (homozygous variant) | 32 (26.89) | 19 (15.83) | (CC vs. TT)0.060.29 (0.06–1.30) |
| 5 Alleles | |||
| C | 170 | 194 | 0.01 |
| T | 68 | 46 | 0.6 (0.37–0.92) |
Also, it was seen that Carriers of Fc¿R1β (T allele) had a more significant risk of developing CAU than those with C allele (P=0.01, OR=0.6, 95% CI=0.37–0.92) as shown in Table 4.
DiscussionWe investigated single SNP of Fc¿R1β gene that might be associated with CSU pathogenesis, further we analyzed the association of total serum IgE levels with the ASST positive CSU patients. In the present study, no significant differences were found in the allele and genotype frequencies of SNP, thereby suggesting that the high affinity receptor gene polymorphisms may not be related to the development of CSU phenotype in our population. The high affinity IgE receptor is responsible for initiating allergic response. The binding of an allergen to the receptor bound IgE leads to mast cell activation and the release of histamine, which are responsible for clinical manifestations of urticaria.17
Polymorphisms of the Fc¿R1β gene have been reported to be associated with the atopy, total serum IgE level, bronchial hyper responsiveness, asthma and the basophilic histamine-releasing activity of asthmatic patients.13–16 Since no previous study has been made on the association of Fc¿R1β gene polymorphisms and urticaria, this study, therefore, is the first of its type to investigate whether there is any significant association between the Fc¿R1β gene polymorphism and the CSU phenotype in a Kashmiri population or not. Also, autoimmunity against the high affinity IgE receptor has been reported in chronic urticaria and CAU.10,11 In the present study, we found no significant associations between the SNPs of Fc¿R1β gene and CSU, however, in this study, serum total IgE levels were found varied to different ranges in CAU patients and most of ASST positive CSU patients were found to have high levels of serum total IgE.
Thus we conclude that there is no statistically significant association between Fc¿R1β gene polymorphism and CSU in Kashmiri population; however, there is a probability of developing CSU in patients carrying Fc¿R1β T allele. Furthermore, serum total IgE levels had no significant association with the development of CAU.
Financial supportThe study was all self funded.
Ethical disclosuresRight to privacy and informed consentThe authors declare that no patient data appear in this article.
Confidentiality of dataThe authors declare that they have followed the protocols of their work center on the publication of patient data.
Protection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Conflict of interestThe authors declared no conflict of interest.
We would like to thank the entire staff of the Immunology & Molecular Medicine Dept., SKIMS for their overwhelming support and help.