Neuropeptide S Receptor (NPSR1) gene has been associated with multiple allergic phenotypes in several patient populations.
ObjectiveWe analysed the effect of the NPSR1 genotypes in the development of asthma, rhinitis, eczema, or food allergy in children randomly receiving either probiotic or placebo treatment.
Methods796 children born to families at high risk for allergic diseases were examined by a paediatrician at the age of three months, six months, two years, and five years. Asthma, rhinitis, eczema, and food allergy were diagnosed according to international guidelines. Treatment with probiotics (double-blinded and placebo controlled) was begun with mothers at 35 weeks of gestation age and continued after the birth of infants up to the age of six months. Association and additive inheritance models were used in genetic analyses.
ResultsDistribution of the hopo546333 was suggestive in the group of patients with atopic eczema at two years. The hopo546333_G was found more often in those with eczema in the placebo group (p=0.048, after Bonferroni correction) and the hopo546333_A was found more often in those with eczema and probiotics compared to those with eczema and placebo treatment. None of the NPSR1 tagging SNPs was associated with asthma, IgE-mediated asthma, or sensitisation. Allergic disease in both parents doubled the risk for IgE-mediated allergic disease (OR 2.1).
ConclusionsThe NPSR1 gene SNP hopo546333 showed a suggestive association for high IgE-associated atopic eczema at two years.
The Neuropeptide S Receptor (NPSR1) gene on chromosome 7 has been previously associated to asthma, high serum total IgE level, and their combination in several populations.1–5 In two large cohorts of children (PARSIFAL and BAMSE), specific NPSR1 haplotypes were associated with atopic sensitisation and asthma,3 but controversial and negative results have also been found for asthma, atopy, and NPSR1 in children.6 Furthermore, an interaction between allergen exposure (farm animal contact) and specific NPSR1 alleles (rs323922 and rs324377) has been reported on allergy outcome in children.7 Other specific NPSR1 SNPs (single nucleotide polymorphisms) (rs324981) have to be reported to associated with bronchial hyperresponsiveness.8
In addition to asthma and allergy, association of NPSR1 SNPs has also been found in other inflammatory diseases such as rheumatoid arthritis, inflammatory bowel disease and intermediate phenotypes of functional gastrointestinal disorders as well as in anxiety disorders.9–11 Correspondingly, expression of NPSR1 has been reported in bronchial epithelium and gastrointestinal tract and in rat brains.11,12 NPSR1 has been reported with several splice variants, promoter polymorphisms affecting its expression on leukocytes and in general it has been found to be expressed in macrophages, neutrophils and intraepithelial lymphocytes.12–14 However, the exact role of NPSR1 in asthma or atopy still remains unsolved.
It has been suggested that probiotics (in detail, see methods) can decrease the risk of onset of allergic rhinitis and asthma by preventing atopic eczema.15,16 Frequencies of allergic and IgE-associated allergic disease did not differ in the probiotic and the placebo groups at the age of five years in previous studies with our population.17 In this same population of children, however, probiotics prevented atopic eczema during the first two years of life, and in addition, they prevented atopic disorders up to the age of five years in children born by caesarean section.18 The principle aim of this study was to determine whether the genetic variants of NPSR1 have an effect on asthma, rhinitis, eczema, or food allergy in early childhood for high-risk families, in addition to the potential effect of immunomodulatory treatment.
MethodsStudy population and treatment protocol with probioticsThe children (N=796) were born between November 2000 and March 2003 to the families in which at least one parent had physician-diagnosed asthma, allergic rhinitis, or atopic dermatitis in the Helsinki suburban area. In this double-blinded, placebo-controlled study, mothers of the birth cohort were randomised at 35 weeks of gestational age into probiotic and placebo groups. From week 36 of gestation, the mothers of the active group received a capsule containing a combination of probiotics (Lactobacillus rhamnosus GG (ATCC 53103) 5×109 colony-forming units (cfu) (Valio Ltd, Helsinki, Finland), L. rhamnosus LC705 5×109, Bifidobacterium breve Bbi99 5×109, Propionibacterium freudenreichii ssp shermanii JS 2×109cfu) twice daily.18 After birth, the infants were given the same capsule once daily along with 0.8g of prebiotic galacto-oligosaccharides or placebo until the age of six months. The newborns were breast-fed and if breast-feeding was insufficient, normal cow's milk-based formula was used, except in cow's milk allergic infants for whom hypoallergenic formulas were given. The Exclusive breast-feeding was for 2.6 months (SD 1.9) in the probiotic group and 2.4 months (SD 2.0) in the placebo group. Likewise, the total breast-feeding time was 8.6 months (SD 5.4) in the probiotic group and 8.2 months (SD 4.9) in the placebo group.17
Assessment of any allergic diseaseChildren were examined by a paediatrician at the age of three months, six months, two years, and five years.19 Asthma, rhinitis and eczema were diagnosed according to clinical guidelines. Atopic eczema was diagnosed if an itching skin condition was accompanied with dry skin and eczema on typical skin areas.20 Asthma was defined when a child has had wheezing episodes at least twice diagnosed by a physician accompanied by prolonged cough or exercise-induced symptoms or bronchial hyperresponsiveness in impulse oscillometry. Allergic rhinitis was diagnosed based on typical symptoms (nasal discharge, blockage, and sneeze/itch) during the pollen season or recurrently in animal contact accompanied with IgE sensitisation against animal epithelia, pollen, or both.21 Food allergy was diagnosed by an open food challenge.22 Any allergic disease combined with IgE sensitisation was considered IgE-associated. IgE sensitisation was shown by positive skin prick tests (ALK-Abellò, Hørsholm, Denmark or Stallergenes, Antony, France) against milk, egg, wheat, fish, peanut, birch, timothy grass, mugwort, cat and dog, and serum-specific IgE antibodies against common food or inhalant allergens. A child was considered sensitised if he/she had a positive skin prick test reaction (≥3mm than negative control), or any serum-specific IgE>0.7kU/L. As an objective measure of the extent and strength of atopic sensitisation, we used the arithmetic SPT sum of all measured food and pollen/animal sensitisations at two and five years. Data on safety were obtained from regular questionnaires at 3, 6 and 12 months and thereafter yearly, and at doctor's clinical assessments at three months, six months, two years, and five years. Growth was recorded at every visit.
DNA extractionBlood samples for genotyping studies were taken at the research visits at two or five years. DNA was extracted from the buffy coat and the following SNPs were analysed for the NPSR1 gene: rs323917 (C/G); rs323922 (G/C); rs324377 (C/A); rHopo546333(G/A); rs324396(C/T); and rs740347(G/C).3 Minor allele frequencies were 0.060 (G) (rs323917); 0.48 (C) rs323922; 0.49 (A) rs324377; 0.096 (A) rHopo546333; 0.34 (T) rs324396; and 0.17 (C) rs740347. For six blood samples, the amount of DNA was insufficient to analysis.
The Ethics Committee of the Hospital for Children and Adolescents of Helsinki University approved the study protocol, and informed consent was signed by the parent.
Statistical methodsChi-square or Fisher's exact test was used to compare difference between groups. Odd ratios and their 95% confidence intervals were calculated for asthma, IgE-associated asthma, any allergic disease (asthma, rhinitis, eczema or food allergy), IgE-associated allergic disease, and for the sum of the prick tests by logistic regression analyses. Distribution of SNP alleles was also studied for IgE-associated allergic disease at 0–5 years and IgE-associated atopic eczema at two years. In logistic regression, the following variables were used: cat or dog at home, respiratory infections at the age of 0–5 years, and use of antibiotics (0–2 years or 3–5 years). The six analysed SNPs were used with an additive inheritance model and the analyses were adjusted for gender, both parents’ allergic disease, parents’ smoking, and placebo or active treatment. In the additive model, alleles at different loci either add a fixed amount to the phenotype or add nothing. The greater the number of trait risk alleles inherited, the more persistent the phenotype is. Controls were defined as those without asthma, without atopic disease, or those with the prick test sum<3mm. Analyses were performed using the SPSS 19.0 software package (SPSS Inc., Chicago, IL, USA), the SAS statistical software, version 9.2 (SAS Institute, Inc., Cary, NC, USA), and PLINK.23 A p-value of less than 0.05 was considered significant.
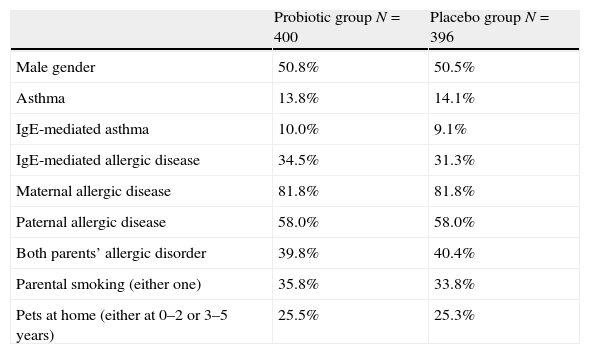
ResultsWe studied the potential effect of the variants of the NPSR1 gene on the development of allergic disease up to the age of five years using association and additive inheritance models. The children had received either probiotic or placebo treatment until the age of six months. There were no differences between probiotic and placebo groups in gender, IgE-associated asthma or IgE-associated allergic disease at the age of five years, parental allergic diseases, parental smoking habits or pets at home (Table 1).
Clinical characteristics of the probiotic and the placebo groups.
| Probiotic group N=400 | Placebo group N=396 | |
| Male gender | 50.8% | 50.5% |
| Asthma | 13.8% | 14.1% |
| IgE-mediated asthma | 10.0% | 9.1% |
| IgE-mediated allergic disease | 34.5% | 31.3% |
| Maternal allergic disease | 81.8% | 81.8% |
| Paternal allergic disease | 58.0% | 58.0% |
| Both parents’ allergic disorder | 39.8% | 40.4% |
| Parental smoking (either one) | 35.8% | 33.8% |
| Pets at home (either at 0–2 or 3–5 years) | 25.5% | 25.3% |
No statistical differences between the study groups (Pearson's chi-square test).
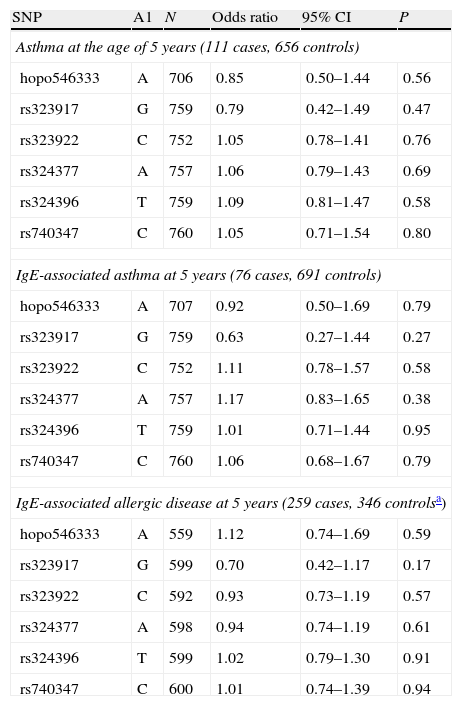
None of the studied chromosome 7 NPSR1 gene SNPs was associated with asthma or IgE-mediated asthma in the additive inheritance model (Table 2). Using the same model we also studied the effect of the genetic variants in the development of sensitisation (the skin prick test sum>3mm) without finding any significant associations (data not shown). IgE-associated allergic disease was not associated with the used NPSR1 gene SNPs when those with no allergic disease and no IgE sensitisation were used as controls (the strictest definition for controls) in the additive inheritance models.
Association of NPSR1 SNP alleles with additive inheritance model for asthma (at the age of 5 years); IgE-associated asthma (at 5 years); and IgE-associated allergic disease (at 5 years).
| SNP | A1 | N | Odds ratio | 95% CI | P |
| Asthma at the age of 5 years (111 cases, 656 controls) | |||||
| hopo546333 | A | 706 | 0.85 | 0.50–1.44 | 0.56 |
| rs323917 | G | 759 | 0.79 | 0.42–1.49 | 0.47 |
| rs323922 | C | 752 | 1.05 | 0.78–1.41 | 0.76 |
| rs324377 | A | 757 | 1.06 | 0.79–1.43 | 0.69 |
| rs324396 | T | 759 | 1.09 | 0.81–1.47 | 0.58 |
| rs740347 | C | 760 | 1.05 | 0.71–1.54 | 0.80 |
| IgE-associated asthma at 5 years (76 cases, 691 controls) | |||||
| hopo546333 | A | 707 | 0.92 | 0.50–1.69 | 0.79 |
| rs323917 | G | 759 | 0.63 | 0.27–1.44 | 0.27 |
| rs323922 | C | 752 | 1.11 | 0.78–1.57 | 0.58 |
| rs324377 | A | 757 | 1.17 | 0.83–1.65 | 0.38 |
| rs324396 | T | 759 | 1.01 | 0.71–1.44 | 0.95 |
| rs740347 | C | 760 | 1.06 | 0.68–1.67 | 0.79 |
| IgE-associated allergic disease at 5 years (259 cases, 346 controlsa) | |||||
| hopo546333 | A | 559 | 1.12 | 0.74–1.69 | 0.59 |
| rs323917 | G | 599 | 0.70 | 0.42–1.17 | 0.17 |
| rs323922 | C | 592 | 0.93 | 0.73–1.19 | 0.57 |
| rs324377 | A | 598 | 0.94 | 0.74–1.19 | 0.61 |
| rs324396 | T | 599 | 1.02 | 0.79–1.30 | 0.91 |
| rs740347 | C | 600 | 1.01 | 0.74–1.39 | 0.94 |
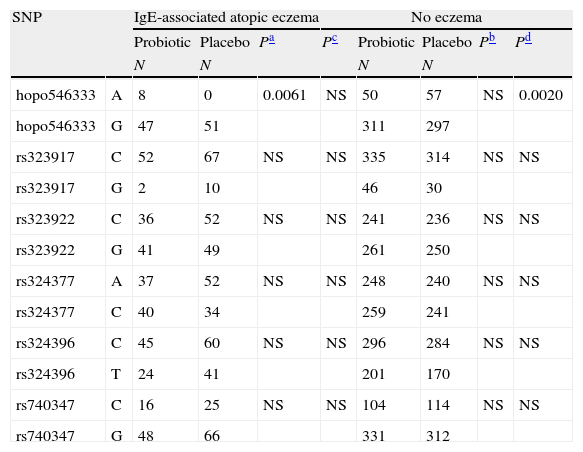
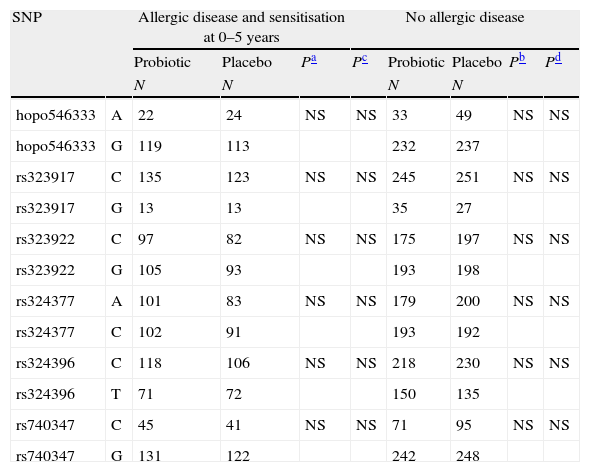
Further, we studied the distribution of SNP alleles separately in the probiotic and placebo groups in high IgE-associated atopic eczema at two years and in allergic disease associated with sensitisation at 0–5 years (Tables 3 and 4, respectively). The distribution of the SNP hopo546333 alleles showed a suggestive association for high IgE-associated atopic eczema (Table 3) at the age of two years. The hopo546333_G was found more often in children with eczema and placebo treatment compared to those without eczema and placebo treatment (p=0.0020). Similarly, the hopo546333_A was found more often in children with eczema and probiotics compared to those with eczema and placebo treatment (p=0.0061). The association of hopo546333_G with atopic eczema in the placebo treatment group remained even after Bonferroni correction for multiple testing (p=0.048). There was no significant association of the SNPs to the studied outcomes when the studied individuals were grouped by caesarean section (data not shown).
Distribution of the NPSR1 gene alleles for high IgE-associated atopic eczema at 2 years of age. The results are presented in probiotic and placebo groups separately.
| SNP | IgE-associated atopic eczema | No eczema | |||||||
| Probiotic | Placebo | Pa | Pc | Probiotic | Placebo | Pb | Pd | ||
| N | N | N | N | ||||||
| hopo546333 | A | 8 | 0 | 0.0061 | NS | 50 | 57 | NS | 0.0020 |
| hopo546333 | G | 47 | 51 | 311 | 297 | ||||
| rs323917 | C | 52 | 67 | NS | NS | 335 | 314 | NS | NS |
| rs323917 | G | 2 | 10 | 46 | 30 | ||||
| rs323922 | C | 36 | 52 | NS | NS | 241 | 236 | NS | NS |
| rs323922 | G | 41 | 49 | 261 | 250 | ||||
| rs324377 | A | 37 | 52 | NS | NS | 248 | 240 | NS | NS |
| rs324377 | C | 40 | 34 | 259 | 241 | ||||
| rs324396 | C | 45 | 60 | NS | NS | 296 | 284 | NS | NS |
| rs324396 | T | 24 | 41 | 201 | 170 | ||||
| rs740347 | C | 16 | 25 | NS | NS | 104 | 114 | NS | NS |
| rs740347 | G | 48 | 66 | 331 | 312 | ||||
Distribution of the NPSR1 gene alleles for allergic disease associated with sensitisation at 0–5 years of age. The results are presented in probiotic and placebo groups separately.
| SNP | Allergic disease and sensitisation at 0–5 years | No allergic disease | |||||||
| Probiotic | Placebo | Pa | Pc | Probiotic | Placebo | Pb | Pd | ||
| N | N | N | N | ||||||
| hopo546333 | A | 22 | 24 | NS | NS | 33 | 49 | NS | NS |
| hopo546333 | G | 119 | 113 | 232 | 237 | ||||
| rs323917 | C | 135 | 123 | NS | NS | 245 | 251 | NS | NS |
| rs323917 | G | 13 | 13 | 35 | 27 | ||||
| rs323922 | C | 97 | 82 | NS | NS | 175 | 197 | NS | NS |
| rs323922 | G | 105 | 93 | 193 | 198 | ||||
| rs324377 | A | 101 | 83 | NS | NS | 179 | 200 | NS | NS |
| rs324377 | C | 102 | 91 | 193 | 192 | ||||
| rs324396 | C | 118 | 106 | NS | NS | 218 | 230 | NS | NS |
| rs324396 | T | 71 | 72 | 150 | 135 | ||||
| rs740347 | C | 45 | 41 | NS | NS | 71 | 95 | NS | NS |
| rs740347 | G | 131 | 122 | 242 | 248 | ||||
In the sub-cohort participating in the NPSR1 genotyping study (N=790 offspring), allergic disease of both parents doubled the risk for IgE-associated allergic disease (OR 2.1, 95% CI 1.47–3.10). Respiratory infections (OR 1.04, 95% CI 1.008–1.07) and use of antibiotics (at 3–5 years) (OR 1.1, 95% CI 1.045–1.17) increased the risk of allergic diseases slightly but significantly. In contrast, pets at home were a protecting factor for IgE-mediated allergic disease (OR 0.58, 95% CI 0.36–0.91). Similarly, for IgE-associated asthma, pets at home (at the age of 3–5 years) were a protecting factor (OR 0.19, 95% CI 0.068–0.54). Again, respiratory infections and use of antibiotics (use of antibiotics was analysed in two parts: at 0–2 years of age and at 3–5 years of age) slightly increased the risk of IgE-associated asthma (1.1, 95% CI 1.038–1.13, 1.2, 95% CI 1.02–1.31 and 1.1, 95% CI 1.032–1.23, respectively). No difference in neonatal morbidity, feeding-related behaviours or serious adverse events between the study groups could be detected. Children in both groups grew normally.
DiscussionIn this first prospective placebo-controlled pharmacogenetic study of the effect of probiotics together with prebiotic oligosaccharide treatment in the prevention of allergic diseases in early childhood, the SNP hopo546333 of the NPSR1 gene showed a suggestive association for high IgE-associated atopic eczema at two years. According to these results the SNPs hopo546333_G allele was a risk factor for high IgE-associated atopic eczema at the age of two years if there was no probiotic treatment. The hopo546333_A allele was a weak risk for high IgE-associated atopic eczema at the age of two years if the child had received probiotic treatment. These results point to an interaction of probiotics and genetic effect. In the additive inheritance model analyses with asthma or IgE-associated asthma at the age of 0–5 years, IgE-associated allergic disease at the age of five years or with the sum of positive skin prick test results, the analysis results were negative.
Our results with the hopo546333 were suggestive for high IgE-associated atopic eczema at the age of two years and also pointed to a gene and exposure interaction. Previously, SNP546333 had been found to associate with farm animal contact, physician-diagnosed-asthma, and atopic asthma with disease onset under 15 years.7 Likewise, NPSR1 SNPs rs323922 and rs324377 have been reported to associate with current farm animal contact and allergic symptoms of asthma and rhinoconjunctivitis among 3113 children aged 5–13 years.7 The whole genetic region of NPSR1 (or GPRA) has been associated with asthma and increased serum total IgE level in the Finnish population, thus representing a region of interest both for our population and for our studied allergic phenotypes.1 None of the studied SNPs modified the effect of probiotics in allergic disease and sensitisation at five years (Table 4). The results may differ from that previously reported since our study population was smaller and younger, and the actual studied intervention (exposure) was probiotics.
When individuals were grouped by caesarean section, we could not find an association between the studied SNPs and high IgE-associated allergic disease. This is most likely because of the small number of individuals when either caesarean-delivered or normally delivered study objects with probiotic/placebo treatment are further divided into different SNP allele categories. Previously, probiotics have shown a protective effect on atopic disorders up to five years in caesarean-delivered children in this same population.18 Duration of breast-feeding did not differ between the probiotic and placebo groups and was not taken into these analyses, which may be considered as a limitation of the study.17
We have chosen any serum-specific IgE>0.7kU/L as a stricter and more solid marker of IgE-mediated phenotype.17,18 Using 0.7kU/L instead of 0.35kU/L not only decreases the number of allergic classified subjects but also increases the possibility of allergy symptoms in these, making the phenotype stricter and stronger. This was different from the study of Melen et al. who chose to use the limit of >0.35kU/L, which might have an impact on the results.3 Other studies have used elevated serum total IgE level for asthma associated phenotype.1,4 In the study of Vergara et al., allele A of Hopo546333 was found to be protective for asthma and allele C of rs740347 for high total IgE level. We used any serum-specific IgE (>0.7kU/L)-associated asthma as a phenotype for IgE-associated asthma.
These results do support a role for the SNP hopo546333 of the NPSR1 gene to lend a minor effect on the development of high IgE-associated atopic eczema at the age of two years in this population of children. The results were only suggestive, which is not entirely unexpected because variable results have been obtained in different studies of similar size, some supporting and some not supporting an association of NPSR1 with asthma. Inconsistent results have been obtained in both child and adult studies. Given the low power of any individual study,24 results with a borderline statistical significance should thus be interpreted with caution.
The rationale for the use of this specific combination and regimen of probiotics and prebiotic was based on previous demonstrated positive effects of the chosen strains in in vitro studies, and by combining strains a stronger effect was sought.25–32 Previously, with the same combination of probiotics as used here, secretion of IL10 was shown as the evidence for anti-inflammatory activity of chosen bacteria.28 Here, using other than study probiotics was forbidden during the intervention period until the age of two years and faecal counts of supplemented probiotic strains were assessed at six and 24 months. Use of non-study probiotics was continuously recorded and if reported use was daily or >2 days a week, it was adjusted for in the outcome measures.18 At three and six months, the probiotic group was significantly more often colonised with lactobacilli and propionibacteria than the placebo group.17 A longer supplementation to the infants and changing the probiotic bacteria strains might have resulted in stronger influences on immune responses and on allergy development.33–35
It is possible that the strains counteract with each other.27 Until now, the strongest effect on preventing atopic eczema has been shown with Lactobacillus rhamnosus.36 On the other hand, mixtures of bacteria have been more effective in stimulating the immune system than one single strain only.25,26 In addition, despite the promising preliminary findings of a probiotic treatment effect or preventive effect on allergy outcomes, the probiotics have not yet yielded a steady position in clinical treatment options.33,34 The evidence regarding eczema prevention is rather good,37,38 but the results regarding effects on respiratory allergies are only beginning to be understood with some positive studies emerging.33,34 Here, the selection of probiotic bacteria was done using the scientific evidence available at the time of the start of the study. The selection included also the Lactobacillus rhamnosus strain which had been shown to adhere to the intestinal mucosa and colonise well, and remain in the gastrointestinal tract. They had all been used with good safety record by the dairy industry.29,30 The strain had also been used in the treatment of atopic children with positive results.31,32
Here, in these almost 800 children born in high-risk families, the allergic disease of both parents increased the risk for IgE-associated allergic disease with OR 2.1. Respiratory infections and the use of antibiotics had an effect of borderline statistical significance on the risk of allergic disease. Having a dog or a cat at home decreased the risk of allergic disease or IgE-associated asthma.39 Frequent respiratory infections might not only be a precursor of developing asthma but also a risk factor for asthma.40 Likewise, pet/allergen exposure and development of allergic disease is complex and controversial results have been reported.41,42
The strength of this investigation is that this study population has been thoroughly examined with the double-blind, placebo-controlled setting with probiotics. Thus, the diagnoses of asthma, atopic eczema, food allergy, or rhinitis have been made using uniform and internationally accepted guidelines.20–22 In addition, the follow-up of the children has been prospective. A limitation of the study is that there is no other sub-phenotype for asthma other than IgE-associated and non-IgE-associated asthma, since the diagnosis of asthma in small children is based on the history of symptoms and clinical findings, although done according to international clinical guidelines. The asthmatic phenotype may differ in children compared to adults since part of the children are cured from asthmatic symptoms when growing up and part of asthmatics develop asthma rather late in adulthood. Since the probiotic treatment was administered only for the first six months of the infants, no further analyses were done extending further than five years of age. In addition, only one gene region was covered with a relatively few SNPs and all the other immune response genes were omitted.43 Further, we could not take into analyses of DNA methylation pattern which has also been reported to have an effect on the NPSR1 region and asthma and allergy outcomes.44
ConclusionThe preliminary effect of the hopo546333 of the NPSR1 gene was shown for high IgE-associated atopic eczema at the age of two years and the results were suggestive of the SNP and exposure (probiotics) interaction. The results are also in line with the total hereditary effect seen in the study population where allergic disease of both parents doubled the risk for IgE-mediated allergic disease (OR 2.1).
Ethical disclosuresPatients’ data protectionConfidentiality of dataThe authors declare that they have followed the protocols of their work centre on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
FundingThis work was supported by the Finnish Allergy Foundation.
Conflict of interestThe authors declare that they have no relevant conflict of interest.
Clinical trials number: NCT00298337.