This study was performed to evaluate association of gene polymorphisms among proinflammatory cytokines and susceptibility to chronic idiopathic urticaria (CIU).
MethodsNinety patients with prolonged urticaria more than 6 weeks were included as case group. Single nucleotide polymorphisms (SNPs) of IL-6 (G/C −174, G/A nt565) and TNF-α (G/A −308, G/A −238) were evaluated, using polymerase chain reaction (PCR); and the results were compared to the control group.
ResultsG allele was significantly higher in the patients at locus of −238 of promoter of TNF-α gene (p<0.001). Frequency of following genotypes were significantly lower in patients with CIU, compared to controls: AG at −308 and GA at −238 of TNF-α gene (p<0.05 and p<0.001, respectively), CG at −174 and GG at +565 of IL-6 gene (p<0.05). Additionally, following genotypes were more common among patients with CIU: GG at −308 and −238 of TNF-α gene (p<0.05 and p<0.001, respectively), GG at −174 and GA at +565 of IL-6 gene (p<0.05).
ConclusionsPro-inflammatory cytokine gene polymorphisms can affect susceptibility to CIU. TNF-α promoter polymorphisms as well as IL-6 gene polymorphisms are associated with CIU.
Chronic urticaria (CU) is defined as pruritic erythematous, wheals lasting more than 6 weeks. However, it is a transitory condition with 1–5 year duration that rarely will be lasted for decades, it impacts the quality of life and usually does not respond to conventional antihistamine therapies with more than 50% failure.1 Mast cells and basophils are the key players in promotion of the skin manifestations in chronic urticaria but data regarding the reasons of their activation and degranulation in these patients is very poor.2,3 Chronic idiopathic urticaria (CIU) is referred to the situation when no cause was detected for the mast cell or basophil degranulation.1
Proinflammatory cytokines including Tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) are implicated in initiation and progression of inflammation in autoimmune and infectious disease and also CU.3–6 They activate inflammatory cells, regulate cell differentiation and apoptosis and will be produced by activated inflammatory cells and macrophages resembling a vicious cycle. Recently, it has been postulated that TNF-α inhibitors may be more beneficial in patients with refractory CIU to other immunomodulators.7 TNF-α was found to be upregulated on endothelial and perivascular cells of the upper dermis in CIU8; this upregulation could be due to environmental stimulus like infections (CU tends to be worsened following infections) or genetic background of the patients (promoter or coding strand allelic polymorphisms). It has already been established that proinflammatory cytokine production is affected by specific gene polymorphisms,9 while association of these cytokines single nucleotide polymorphisms (SNPs) have been studied in a number of immunological diseases.10–14 Hence, the gene polymorphisms may be an important actor in CIU pathogenesis, in which environmental trigger is unknown.
With respect to the immunologic dysregulation in CIU supported by observation of high serum levels of IL-6 and TNF-α that induce basophil activation, histamine release and leukotriene production,15,16 polymorphisms of promoter or coding sequences of proinflammatory cytokines have been studies in a number of patients with CIU.
Patients and methodsStudy designThis study comprised 90 patients with CIU (75 females), who referred to the Children's Medical Center, the Pediatrics Center of Excellence in Tehran, Iran. A detailed history was taken from all participants; their medical records were reviewed to ascertain their eligibility for inclusion to the study. Patients with established diagnosis of CIU according to standard international guidelines17,18 were enrolled from the outpatient clinic. The diagnosis of CIU was considered for the patient when the wheals lasting for 6 weeks or longer with occurrences of at least 2 times a week and its underlying cause remained unclear despite the appropriate investigations. Patients with physical urticaria, food/medication induced allergy or urticarial vasculitis were excluded. Other exclusion criteria were urticaria lesions due to infections, environmental agents, stress or cholinergic stimulation and heat/cold induced urticaria, dermatographic urticaria and pressure induced urticaria. To correctly exclude aforementioned etiologies, the onset and duration of lesions, characteristics of lesions, distribution of lesions, and history of any associated disease or allergies, drug history and complement deficiencies were considered. Specific laboratory tests were performed to approve the suspicious diagnosis, for instance C3, C4, CH50 and C1 inhibitor (C1-INH) for angioedema. One hundred and thirty nine healthy individuals who had no evidence of allergic, autoimmune or any other systemic disease were also considered as control group.19 The local ethic committee and appropriate institutional review board approved the study; all the participants signed the informed consent forms prior to enrollment.
Sampling and genotypingAmount of 5 milliliters (ml) of whole blood was drawn from all participants and kept with Ethylene-diamine-tetra-acetic acid (EDTA) at −20°C until investigation. DNA was extracted from nucleated cells using phenol-chloroform as described previously.20 For SNP detection, polymerase chain reaction with the sequence specific primers (PCR-SSP) assay (PCR-SSP kit, Heidelberg University, Heidelberg, Germany) was employed. Extracted DNA was amplified using a PCR Techne Flexigene apparatus (Rosche, Cambridge, UK), as explained before.19 PCR products were observed by 2% agarose gel electrophoresis and a picture was taken after visualization with a UV transilluminator. Following single nucleotide gene polymorphisms were evaluated: IL-6 (G/C −174, G/A nt565) and TNF-α (G/A −308, G/A −238). The investigator responsible for genotype analysis was blinded to the study.
Statistical analysisThe descriptive variables were expressed as frequency or median with range. Allele and genotype frequencies were determined by direct gene counting and analyzed by chi-square test. A little discrepancy from Hardy–Weinberg equilibrium was only observed in TNF-α (A/G, −238) and IL-6 (C/G, 174) alleles that was not statistically significant (p=0.992, p=0.833). Hence, all of the alleles evaluated in this study comply with Hardy-Weinberg equilibrium. Odds ratios (OR) and 95% confidence intervals (CI) were estimated. All the tests were two-sided and the probability of less than 0.05 was considered as statistically significant. The analyses were performed using the Epi Info statistical software (version 6.2, World Health Organization, Geneva, Switzerland).
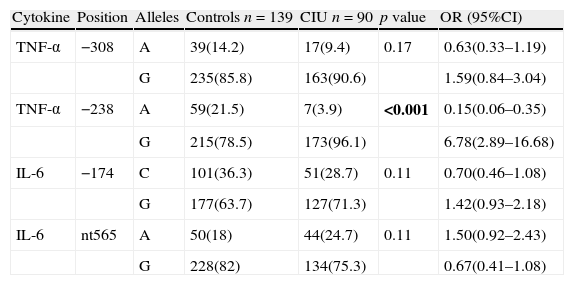
ResultsAllele frequenciesSNP analysis between patients with CIU and controls revealed no difference in IL6 (G/C −174, G/A nt565). However, frequency of alleles A/G at position −238 in the promoter of TNF-α gene was significantly different between patients with CIU and controls (G to A allele ratio was 24.1 vs. 3.6 in CIU vs. controls, respectively). Despite of increased G allele at position −308 in patients with CIU compared to controls, it did not achieve significance. Detail allele frequencies for both patient and control groups have been depicted in Table 1.
Allele frequencies of TNF-α and IL-6 genes in patients with urticarial and controls.
| Cytokine | Position | Alleles | Controls n=139 | CIU n=90 | p value | OR (95%CI) |
| TNF-α | −308 | A | 39(14.2) | 17(9.4) | 0.17 | 0.63(0.33–1.19) |
| G | 235(85.8) | 163(90.6) | 1.59(0.84–3.04) | |||
| TNF-α | −238 | A | 59(21.5) | 7(3.9) | <0.001 | 0.15(0.06–0.35) |
| G | 215(78.5) | 173(96.1) | 6.78(2.89–16.68) | |||
| IL-6 | −174 | C | 101(36.3) | 51(28.7) | 0.11 | 0.70(0.46–1.08) |
| G | 177(63.7) | 127(71.3) | 1.42(0.93–2.18) | |||
| IL-6 | nt565 | A | 50(18) | 44(24.7) | 0.11 | 1.50(0.92–2.43) |
| G | 228(82) | 134(75.3) | 0.67(0.41–1.08) |
n, denotes to number of participants in each group. Bold values signify p<0.05.
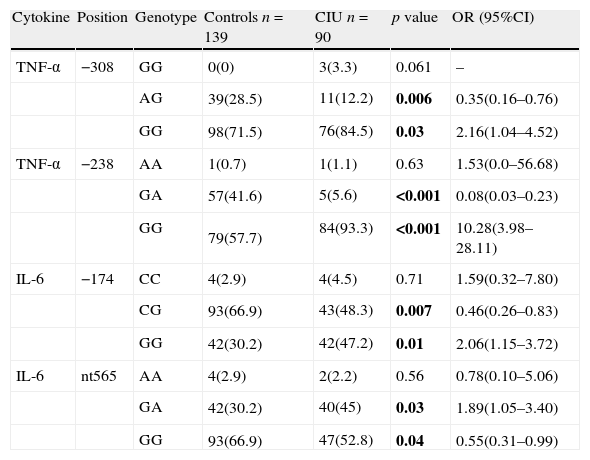
Genotype GG was significantly higher at both positions of −238 and −308 of promoter sequence of TNF-α gene in patients with CIU. Furthermore, heterozygotes for alleles A/G were significantly lower in patients with CIU with respect to controls. In contrast to allele frequencies, GG and GA genotypes were higher and CG and GG genotypes were lower in patients at positions of −174 and +565 of IL-6 gene, respectively. Detail of genotype frequencies have been demonstrated in Table 2.
Genotype frequencies of TNF-α and IL-6 genes in patients with urticarial and controls.
| Cytokine | Position | Genotype | Controls n=139 | CIU n=90 | p value | OR (95%CI) |
| TNF-α | −308 | GG | 0(0) | 3(3.3) | 0.061 | – |
| AG | 39(28.5) | 11(12.2) | 0.006 | 0.35(0.16–0.76) | ||
| GG | 98(71.5) | 76(84.5) | 0.03 | 2.16(1.04–4.52) | ||
| TNF-α | −238 | AA | 1(0.7) | 1(1.1) | 0.63 | 1.53(0.0–56.68) |
| GA | 57(41.6) | 5(5.6) | <0.001 | 0.08(0.03–0.23) | ||
| GG | 79(57.7) | 84(93.3) | <0.001 | 10.28(3.98–28.11) | ||
| IL-6 | −174 | CC | 4(2.9) | 4(4.5) | 0.71 | 1.59(0.32–7.80) |
| CG | 93(66.9) | 43(48.3) | 0.007 | 0.46(0.26–0.83) | ||
| GG | 42(30.2) | 42(47.2) | 0.01 | 2.06(1.15–3.72) | ||
| IL-6 | nt565 | AA | 4(2.9) | 2(2.2) | 0.56 | 0.78(0.10–5.06) |
| GA | 42(30.2) | 40(45) | 0.03 | 1.89(1.05–3.40) | ||
| GG | 93(66.9) | 47(52.8) | 0.04 | 0.55(0.31–0.99) |
n, denotes to number of participants in each group. Bold values signify p<0.05.
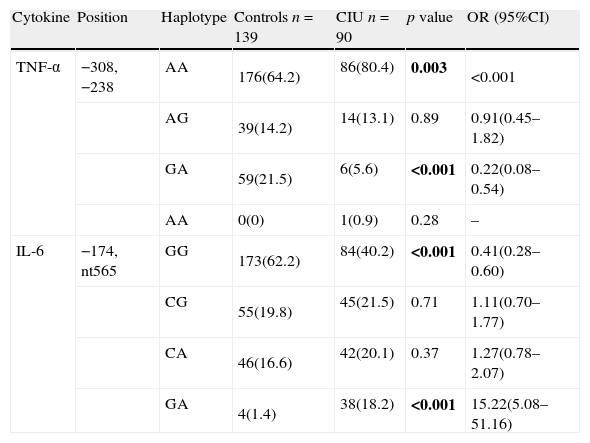
The haplotype GG of TNF-α at positions −308, −238 in the patients with CIU was significantly higher than the control group, while GA haplotype had significantly lower frequency in the patient group. Frequency of IL-6 −174, nt565 haplotype in the patient group was significantly lower than the controls (Table 3).
Haplotype frequencies of TNF-α and IL-6 genes in patients with urticarial and controls.
| Cytokine | Position | Haplotype | Controls n=139 | CIU n=90 | p value | OR (95%CI) |
| TNF-α | −308, −238 | AA | 176(64.2) | 86(80.4) | 0.003 | <0.001 |
| AG | 39(14.2) | 14(13.1) | 0.89 | 0.91(0.45–1.82) | ||
| GA | 59(21.5) | 6(5.6) | <0.001 | 0.22(0.08–0.54) | ||
| AA | 0(0) | 1(0.9) | 0.28 | – | ||
| IL-6 | −174, nt565 | GG | 173(62.2) | 84(40.2) | <0.001 | 0.41(0.28–0.60) |
| CG | 55(19.8) | 45(21.5) | 0.71 | 1.11(0.70–1.77) | ||
| CA | 46(16.6) | 42(20.1) | 0.37 | 1.27(0.78–2.07) | ||
| GA | 4(1.4) | 38(18.2) | <0.001 | 15.22(5.08–51.16) |
n, denotes to number of participants in each group. Bold values signify p<0.05.
This is the unique study evaluated proinflammatory cytokine gene polymorphisms among patients with CIU. The results suggested that TNF-α promoter polymorphisms as well as IL-6 gene polymorphisms are associated with CIU. However, the exact mechanism of gene polymorphism effect on up/down regulation of genes is unclear, but it can affect both transcription and translation activities make the carrier prone/resistant to urticaria.9 There are several steps to regulate the gene expression but the most important one is at initiation of transcription and RNA polymerase binding. Polymorphisms in promoter region may cause up/down regulation of target gene via change in affinity of DNA binding proteins; it is also suggested that some repressor proteins only bind to one allele due to different 3-dimensional configuration of DNA at polymorphic site.21 Moreover, not only un-translated regions (UTRs) may affect the stability of messenger RNA (mRNA), but also coding sequences could do similar to what Kim et al. demonstrated that Histamine N-methyltransferase 939A>G polymorphism lowers the corresponding mRNA stability with deceased enzymatic activity and increased histamine levels in patients with aspirin intolerant chronic urticarial.22 Accordingly, gene polymorphisms can be regarded as a useful genetic marker that may determine predisposition or resistance to CIU.
TNF-α is the pivotal proinflammatory cytokine in induction of inflammation, apoptotic cell death and as the name specified inhibition of tumorigenesis and can induce release of other proinflammatory cytokines. Previously, the link between susceptibility to aspirin induced-urticaria (AIU) and polymorphisms at −1031T>C and −863C>A, but not −238G>A and −308G>A of the promoter of TNF-α was shown by Choi et al. in the Korean patients.23 Furthermore, Davis et al. demonstrated that TNF2 allele carriage is associated with increased erythema in patients with irritant contact dermatitis.24 On the other hand, the association of TNF-α polymorphism at −238 and −308 (A allele) with contact dermatitis was demonstrated in a sample of 86 patients in Germany25 and a sample of 197 patients in the Netherlands,26 but no association was found in Chinese patients (n=94).27 Overall, it seems that polymorphisms at promoter region of TNF-α gene may predispose the carrier to pruritic erythematous chronic inflammatory skin disease.
We found that polymorphic site of −238 in the promoter of TNF-α was associated with susceptibility to CIU and was occupied more with G allele in patients with CIU (p<0.001). This was further supported by genotype analysis in which GG was the most frequent genotype in CIU patients and was significantly higher compared to controls (frequency of 93.3% and 84.5% at loci −238 and −308, respectively). It is demonstrated that substitution of G to A at loci −238 and −308 (TNF2) of the promoter of TNF-α strongly increases the transcription activity.9 Accordingly, higher frequency of A allele at aforementioned loci was expected in CIU patients, but inversely this study found higher frequency for G allele. The observed allele and genotype frequencies cannot be due to the ethnic differences as this research team previously demonstrated no significant difference in allele polymorphisms at −238/−308 of TNF-α promoter among Iranian and Caucasians from Italy, Greek and England.19 One can be suggested that in CIU the observed polymorphisms may induce upregulation of TNF-α. Kroeger et al. demonstrated that transcriptional activity of TNF2 allele is cell type specific so depending on the pathology behind each inflammatory disease increased expression of TNF-α may be attributed to TNF1/TNF2 allele.28 It cannot be claimed that TNF1 and other more frequent polymorphisms observed in this study can induce upregulation of TNF-α, as we did not evaluate gene expression or cytokine levels and to our knowledge there is not such a study in Iranian patients with CIU. Nevertheless, TNF-α is widely accepted to be upregulated not only in skin microenvironment (both affected and uninvolved regions), but also in sera of patients with CIU and different ethnicities (Italian,29 German,8 Brazilian).15
IL-6 is a marker of systemic inflammation and a strong stimulator of synthesis of acute phase reactant proteins. It is believed that IL-6 concentration is increased in CIU and was suggested to be implicated in neuroendocrine disturbance found in CIU.29 Increase in serum IL-6 level in patients with CIU is correlated with disease activity and may be elicited by both environmental triggers (i.e. infections) or genetic predispositions.30 G to C substitution at −174 of IL-6 is associated with 2 fold increase in in vivo transcriptional activity. In this study, despite the increased frequency of G allele in CIU patients it did not attain significance. However, genotype analysis was significantly different between CIU patients and controls in which CC:CG:GG frequencies were 4.5%:48.3%:47.2% in patients with CIU and 2.9%:66.9%:30.2% in controls. The most frequent genotype was CG, but in patients with CIU it was decreased and GG genotype was increased significantly (p<0.05). Recently, Keyserova et al. demonstrated a significant association between −174 (C/G, G/G) and nt565 (A/G, G/G) and susceptibility to atopic dermatitis,31 but this study is the first report indicating the role of IL-6 gene polymorphism in susceptibility to CIU.
One of limitations that should be acknowledged is that the level of proinflammatory cytokines in sera or skin biopsies as the final phenotype was not measured. Furthermore, the transcription modification by polymorphisms could be assessed using luciferase receptor assay that was not performed in this study. Despite of appropriate design and statistical methods employed in this study, another limitation was the possibility of the presence of confounding factors due to the poor data upon factors can affect the susceptibility to CIU. Another limitation of this study is the possibility of misdiagnosing of patients with CIU as it is difficult and sometimes impossible to identify the exact etiology of chronic urticaria. With respect to the results of current study and the fact that environmental trigger is unknown in CIU, the role of genetic predisposition seems to be important in susceptibility to CIU. It should be noted that further studies on other populations with different ethnicities are needed to warrant results of this study and to evaluate the genetic markers in CIU and possible association with the course of disease.
Ethical disclosuresProtection of human subjects and animals in researchProtection of human and animal subjects. The authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Patients’ data protectionConfidentiality of data. The authors declare that they have followed the protocols of their work center on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentRight to privacy and informed consent. The authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Conflict of interestThe authors have no conflict of interest to declare.
This study was supported by a grant from Immunology, Asthma and Allergy Research Institute, Tehran University of Medical Sciences.