Background and aim. To identify the geographic distribution of hepatitis C virus (HCV) genotypes and HCV RNA viral load in a large number of HCV-infected carriers in Mexico.
Methods. Patients with chronic hepatitis C (n = 8,802) were studied to identify HCV genotype using an immune line probe assay in samples shown previously to be positive for viral RNA by an RT-PCR test. Baseline HCV RNA was also evaluated.
Results. Genotype 1 accounted for 70.3%, genotype 2 for 21.8%, genotype 3 for 7.2%, genotype 4 for 0.3%, and genotype 5 for 0.1% of all cases; coinfection was present in 0.3%. Overall, Genotype 1 was the most prevalent Genotype. Regionally, genotype 1 occurred more frequently in the North-East, North, and Center-East regions of Mexico; genotype 2 was more prevalent in the South, East, and Peninsula regions; and genotype 3 was more prevalent in the North and North-West regions. Only 22.4% of patients with genotype 1 were classified in the low HCV RNA viral load category, and the distribution of this genotype did not differ significantly between regions.
Conclusion. The prevalence of HCV genotypes and viral load in Mexico was 70.3% for genotype 1, but only 22.4% of these patients had a low HCV viral load. Distribution was not uniform in Mexico, with greater frequency of genotype 2 in South, East and Peninsula Regions and Genotype 3 in North and North-West Regions.
Hepatitis C virus (HCV) infection associated with chronic hepatitis is one of the most prevalent causes of liver-related mortality and morbidity worldwide.1 HCV infection is a risk factor for hepatocellular carcinoma and cirrhosis.2 Patients with posttransfusional hepatitis C might develop cirrhosis after an average of 21 years and hepatocellular carcinoma after an average of 29 years.3 Because the most serious consequences of HCV appear several years after infection acquisition, to improve outcomes and optimize resources, it is important to identify HCV-infected patients who are at high risk for progressive liver diseases and who may benefit from early therapeutic intervention.
HCV is characterized by a high mutation rate and considerable genomic heterogeneity, which have consequences for the effectiveness of treatment.4 Genotype identification is important clinically because genotypes 1 and 4 are more resistant than are genotypes 2 and 3 to Pegylated interferon and Ribavirin combination therapy, and thus require different treatment duration and dose. Current guidelines establish treatment duration for genotypes 1 and 4 for 48 weeks and 24 weeks for genotype 2 and 3 with Pegylated interferon and Ribavirin.5 In contrast, patients infected with genotype 1 HCV but with a low baseline HCV RNA content (< 200,000 IU/mL) are more likely to achieve a sustained virological response (SVR) than are those with an intermediate (200,000-600,000 IU/mL) or high (> 600,000 IU/ mL) viral load when treated with pegylated interferon and ribavirin for 24 weeks.6 There is emerging evidence that patient who achive rapid viral response (RVR) (< 50 IU/mL) whatever the HCV genotype, may shorten treatment duration, On the other hand, patien with genotype 1 who did reduce by 2 logs the viral load at week 12, but who do not achive a viral load of (<50IU/mL) may have a greater chance to succed if treatment duration in extended.7
One important indicator of viral genotype variation is geographic distribution. For instance, the occurrence of a single HCV type represented by several subtypes suggests a long period of endemic infection. Examples include genotype 1 in the Guinea coast of West Africa, genotype 2 in Central and West Africa, genotype 3 in the Northern Indian subcontinent, genotype 4 in Central Africa, and genotype 6 in South-East Asia.4 In other parts of the world, more than one virus type can be present, but each is represented by a few different subtypes. In Northern Europe and North America, genotypes 1a, 1b, 2b, and 3a are common, whereas in Japan only types 1b, 2a, and 2b occur.8 This genotype distribution is consistent with relatively recent and limited introductions from endemic areas. Genotypes 1a and 3a are found among individuals who have drug injection as a risk factor.9 The assumption of a recent dissemination of these genotypes is supported by the low diversity observed within these subtypes.
In Mexico, few large-scale studies have evaluated the geographic distribution of HCV genotypes among chronic HCV-infected patients. To our knowledge, the most important study to date included 1,390 chronic HCV-infected patients. This study concluded that the distribution of HCV genotypes in four geographic areas in Mexico was not homogeneous and that genotype 3 occurs with greater frequency in the northern region.10 To design treatment strategies at the national level, more information about the genotypes and basal viral load is required. The purpose of this study was to describe the geographic distribution of HCV genotypes in eight different regions and to estimate the magnitude of HCV viral load in patients before therapy. The present work included 8,802 chronic HCV-infected patients studied throughout the Mexican Republic.
MethodsBetween the years 2002 and 2008, patients with chronic hepatitis C were identified as candidates for antiviral treatment within different hospitals in Mexico. The patients included in the study provided written informed consent for participation and allowed drawing of a blood sample. Samples were first tested for the presence of viral RNA by reverse transcription-polymerase chain reaction (RT-PCR) testing (Roche Diagnostics Cobas Amplicor and Cobas TaqMan –Basel, Switzerland–). Samples shown to be positive for the presence of HCV by RT-PCR testing were analyzed further to identify the hepatitis C genotype using an immune line probe assay (INNO-LiPA, Bayer Diagnostics –Tarrytown, NY–).
To study the geographic distribution of HCV genotypes, we used a structural variable11 derived from the economic regionalization of Mexico into eight regions, each including different states as follows: North-West (Baja California, Baja California Sur, Nayarit, and Sinaloa y Sonora), North (Coahuila, Chihuahua, Durango, San Luis Potosí, and Zacatecas), North-East (Tamaulipas and Nuevo León), Center-West (Aguascalientes, Colima, Guanajuato, Jalisco, and Michoacán), Center-East (Federal District, Hidalgo, Estado de México, Morelos, Puebla, Querétaro, and Tlaxcala), South (Chiapas, Guerrero, and Oaxaca), East (Tabasco and Veracruz), and Peninsula (Campeche, Quintana Roo, and Yucatán).12
The simple and relative frequencies of HCV genotype distribution were calculated according to their geographic distribution in these eight regions. To evaluate whether the prevalence of HCV genotype differs by geographic region, the magnitude of the association was calculated using odds ratios (ORs) and their corresponding confidence intervals (CIs), and p values. The simple and relative frequencies of HCV viral load were also calculated, and their distribution was stratified into three categories of magnitude: low (< 200,000 IU/mL), intermediate (200,001–600,000 IU/mL), and high (> 600,000 IU/mL), according to the criteria proposed by Jensen, et al.6 The relationship between HCV viral load and sex, and between HCV viral load and genotype were evaluated using the chi-square test and were also stratified by region.
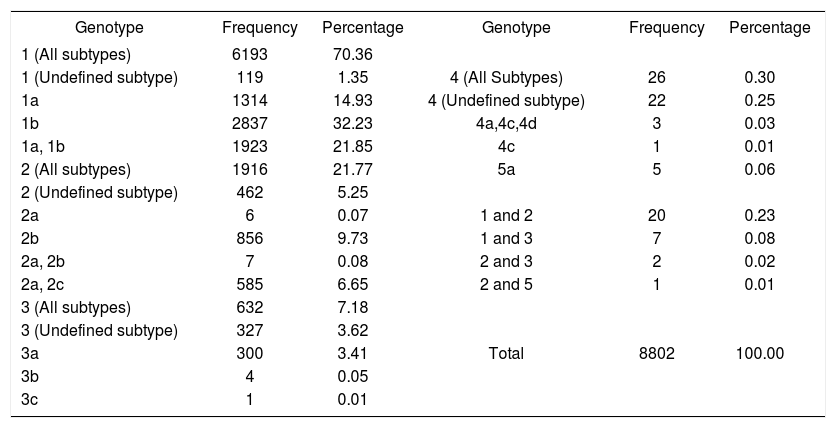
ResultsThe study included 8,802 patients, 57.9% of whom were female. Table 1 shows the distribution of HCV genotype. Genotype 1 was the most frequent and accounted for 70.3% of the cases. Subtype 1b was the most prevalent (32.2% of the cases). Genotype 2 had a frequency of 21.8%, and the most frequent subtype was 2b (9.7%). The frequency of genotype 3 was 7.2%, type 4 was 0.3%, and type 5 was 0.1%. We found the coexistence of different genotypes in the same patient in 30 cases. The most frequent combinations were 1 + 2 (20 cases) and 1 + 3 (seven cases).
HCV genotype distribution among 8,802 patients with chronic hepatitis C.
| Genotype | Frequency | Percentage | Genotype | Frequency | Percentage |
|---|---|---|---|---|---|
| 1 (All subtypes) | 6193 | 70.36 | |||
| 1 (Undefined subtype) | 119 | 1.35 | 4 (All Subtypes) | 26 | 0.30 |
| 1a | 1314 | 14.93 | 4 (Undefined subtype) | 22 | 0.25 |
| 1b | 2837 | 32.23 | 4a,4c,4d | 3 | 0.03 |
| 1a, 1b | 1923 | 21.85 | 4c | 1 | 0.01 |
| 2 (All subtypes) | 1916 | 21.77 | 5a | 5 | 0.06 |
| 2 (Undefined subtype) | 462 | 5.25 | |||
| 2a | 6 | 0.07 | 1 and 2 | 20 | 0.23 |
| 2b | 856 | 9.73 | 1 and 3 | 7 | 0.08 |
| 2a, 2b | 7 | 0.08 | 2 and 3 | 2 | 0.02 |
| 2a, 2c | 585 | 6.65 | 2 and 5 | 1 | 0.01 |
| 3 (All subtypes) | 632 | 7.18 | |||
| 3 (Undefined subtype) | 327 | 3.62 | |||
| 3a | 300 | 3.41 | Total | 8802 | 100.00 |
| 3b | 4 | 0.05 | |||
| 3c | 1 | 0.01 |
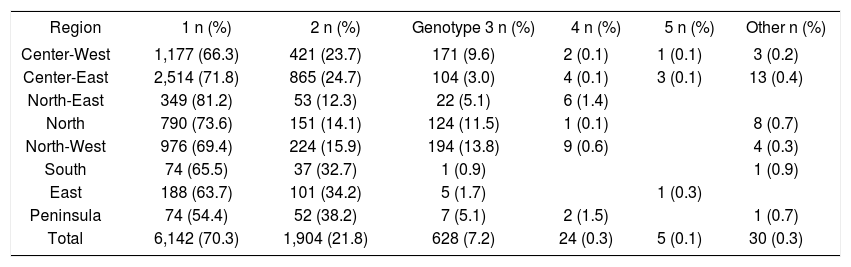
Table 2 shows that genotype 1 was more frequent in the North-East, North, and Center-East regions of Mexico, with prevalence rates of 71.8% to 81.2%, which were above the average. Genotype 2 was more frequent in the South, East, and Peninsula regions, where the prevalence was 32.7% to 38.2%. Genotype 3 prevailed in the North and North-West regions, where the prevalence was 11.5% and 13.8%, respectively. These variations are shown graphically in Figure 1.
HCV genotype distribution in the eight geographic regions of Mexico.
| Region | 1 n (%) | 2 n (%) | Genotype 3 n (%) | 4 n (%) | 5 n (%) | Other n (%) |
|---|---|---|---|---|---|---|
| Center-West | 1,177 (66.3) | 421 (23.7) | 171 (9.6) | 2 (0.1) | 1 (0.1) | 3 (0.2) |
| Center-East | 2,514 (71.8) | 865 (24.7) | 104 (3.0) | 4 (0.1) | 3 (0.1) | 13 (0.4) |
| North-East | 349 (81.2) | 53 (12.3) | 22 (5.1) | 6 (1.4) | ||
| North | 790 (73.6) | 151 (14.1) | 124 (11.5) | 1 (0.1) | 8 (0.7) | |
| North-West | 976 (69.4) | 224 (15.9) | 194 (13.8) | 9 (0.6) | 4 (0.3) | |
| South | 74 (65.5) | 37 (32.7) | 1 (0.9) | 1 (0.9) | ||
| East | 188 (63.7) | 101 (34.2) | 5 (1.7) | 1 (0.3) | ||
| Peninsula | 74 (54.4) | 52 (38.2) | 7 (5.1) | 2 (1.5) | 1 (0.7) | |
| Total | 6,142 (70.3) | 1,904 (21.8) | 628 (7.2) | 24 (0.3) | 5 (0.1) | 30 (0.3) |
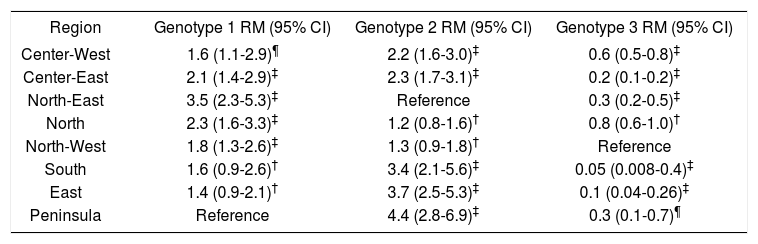
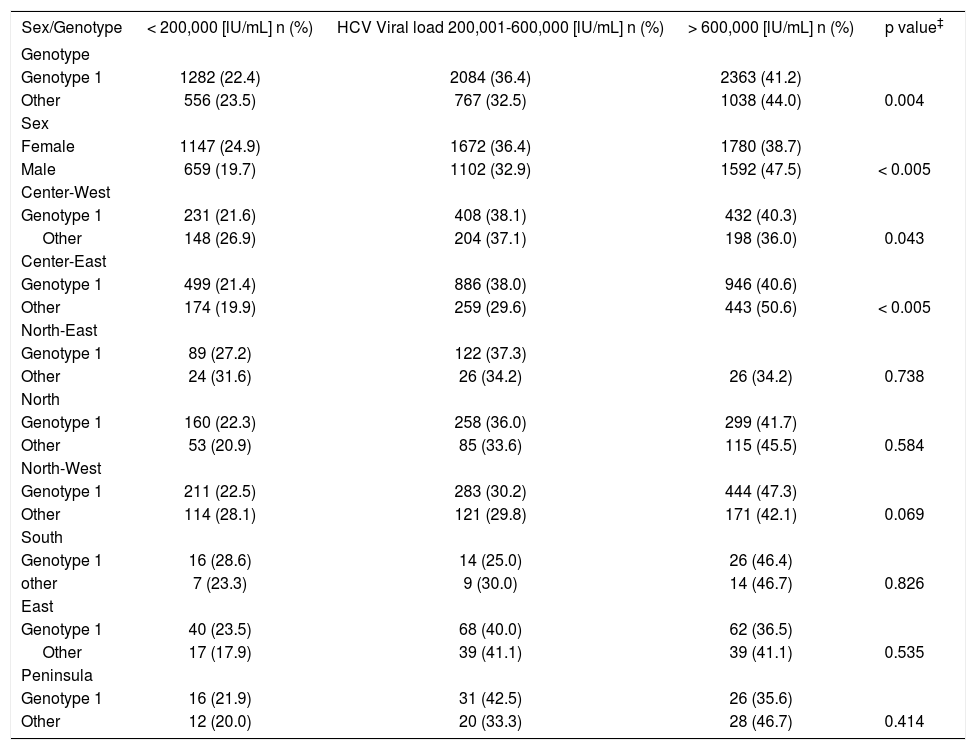
The frequencies of genotypes differed significantly between geographic regions (Table 3). For example, the highest probability of being infected by genotype 1 was 3.5 times higher (95% CI 2.3, 5.3) in the North-East region than in the Peninsula region (Table 3). Similarly, the probability of being infected by genotype 2 was 4.4 times higher (95% CI 2.8, 6.9) in the Peninsula region than in the North-East region, and the probability of being infected with genotype 3was 20 times higher (95% CI 2.5, 125) in the North-West region than in the South region. In this last case, the ORs were calculated using the highest prevalence of HCV genotype 3 in the North-West region as the category of reference. The OR of 20 was estimated by calculating the inverse value of the 0.05 OR.A trend was observed in the frequency of HCV viral load categories according to sex and genotype 1. In females, 24.9%, 36.4%, and 38.7% exhibited low, intermediate, and high viral load, respectively; the values in males were 19.7%, 32.9% and 47.5% (p < 0.005) (Table 4). The distribution of genotype 1 was 22.4%, 36.4%, and 41.2% for low, intermediate, and high viral loads, respectively. The respective distribution for genotypes 2 through 5 was 23.5%, 32.5%, and 44.0% (p < 0.004).
Prevalence of HCV infection by genotypes 1 to 3 in the eight geographic regions of Mexico.
| Region | Genotype 1 RM (95% CI) | Genotype 2 RM (95% CI) | Genotype 3 RM (95% CI) |
|---|---|---|---|
| Center-West | 1.6 (1.1-2.9)¶ | 2.2 (1.6-3.0)‡ | 0.6 (0.5-0.8)‡ |
| Center-East | 2.1 (1.4-2.9)‡ | 2.3 (1.7-3.1)‡ | 0.2 (0.1-0.2)‡ |
| North-East | 3.5 (2.3-5.3)‡ | Reference | 0.3 (0.2-0.5)‡ |
| North | 2.3 (1.6-3.3)‡ | 1.2 (0.8-1.6)† | 0.8 (0.6-1.0)† |
| North-West | 1.8 (1.3-2.6)‡ | 1.3 (0.9-1.8)† | Reference |
| South | 1.6 (0.9-2.6)† | 3.4 (2.1-5.6)‡ | 0.05 (0.008-0.4)‡ |
| East | 1.4 (0.9-2.1)† | 3.7 (2.5-5.3)‡ | 0.1 (0.04-0.26)‡ |
| Peninsula | Reference | 4.4 (2.8-6.9)‡ | 0.3 (0.1-0.7)¶ |
HCV viral load categories and their distribution by sex and genotype, and their stratification by region and genotype.
| Sex/Genotype | < 200,000 [lU/mL] n (%) | HCV Viral load 200,001-600,000 [lU/mL] n (%) | > 600,000 [lU/mL] n (%) | p value‡ |
|---|---|---|---|---|
| Genotype | ||||
| Genotype 1 | 1282 (22.4) | 2084 (36.4) | 2363 (41.2) | |
| Other | 556 (23.5) | 767 (32.5) | 1038 (44.0) | 0.004 |
| Sex | ||||
| Female | 1147 (24.9) | 1672 (36.4) | 1780 (38.7) | |
| Male | 659 (19.7) | 1102 (32.9) | 1592 (47.5) | < 0.005 |
| Center-West | ||||
| Genotype 1 | 231 (21.6) | 408 (38.1) | 432 (40.3) | |
| Other | 148 (26.9) | 204 (37.1) | 198 (36.0) | 0.043 |
| Center-East | ||||
| Genotype 1 | 499 (21.4) | 886 (38.0) | 946 (40.6) | |
| Other | 174 (19.9) | 259 (29.6) | 443 (50.6) | < 0.005 |
| North-East | ||||
| Genotype 1 | 89 (27.2) | 122 (37.3) | ||
| Other | 24 (31.6) | 26 (34.2) | 26 (34.2) | 0.738 |
| North | ||||
| Genotype 1 | 160 (22.3) | 258 (36.0) | 299 (41.7) | |
| Other | 53 (20.9) | 85 (33.6) | 115 (45.5) | 0.584 |
| North-West | ||||
| Genotype 1 | 211 (22.5) | 283 (30.2) | 444 (47.3) | |
| Other | 114 (28.1) | 121 (29.8) | 171 (42.1) | 0.069 |
| South | ||||
| Genotype 1 | 16 (28.6) | 14 (25.0) | 26 (46.4) | |
| other | 7 (23.3) | 9 (30.0) | 14 (46.7) | 0.826 |
| East | ||||
| Genotype 1 | 40 (23.5) | 68 (40.0) | 62 (36.5) | |
| Other | 17 (17.9) | 39 (41.1) | 39 (41.1) | 0.535 |
| Peninsula | ||||
| Genotype 1 | 16 (21.9) | 31 (42.5) | 26 (35.6) | |
| Other | 12 (20.0) | 20 (33.3) | 28 (46.7) | 0.414 |
*p value by chi-square test.
In recent years, several studies have been conducted in Mexico to determine the frequencies of HCV genotypes among different populations in Mexico.13-15 These studies included populations with very low prevalence of antibodies against HCV. As a result, these studies have limitations for estimating both the frequency and geographic distribution of HCV genotypes among chronic carrier persons. Rivas-Estilla and colleagues16 studied 147 patients with anti-HCV antibodies and detectable HCV RNA in a university hospital in the North-East of Mexico. The HCV genotype found most frequently was genotype 1 (73%) and subtype 1b was the most prevalent (37.4%). Two recent reviews of the prevalence of hepatitis C virus in the Mexican population reported that genotype 1 is the most prevalent, with frequencies of 30% to 87.5%17 and 63% to 70%.18 In both reviews, subtype 1b was the most frequent and its frequency ranged between 11.9% and 61.9%(16) and 21% and 47%.18
The most extensive studies of HCV genotype distribution in Mexico were reported by Sánchez-Ávila colleagues10 and our current study. These two studies support the previous proposal that genotype 1 and subtype 1b are the most frequent in chronic HCV carriers in the Mexican population. Our prevalence rates are similar to those reported by Sánchez-Ávila et al.10 For example, Sánchez-Ávila et al reported an HCV genotype 1 frequency of 68.9% and we found a frequency of 70.3% of all cases studied; the respective frequencies of subtype 1b were 33.8% and 32.2%. The second most frequent HCV genotype was genotype 2. The prevalence rates of genotype 2 were 21.4% in the first study10 and 21.8% in our study, and the most frequent subtype was 2b (7% in the first study and 9.7% in our study). The prevalence rates of genotypes 3, 4, and 5 were 9.2%, 0.36%, and 0.07%, respectively, in the earlier study and 7.2%, 0.3%, and 0.1% in our study.
We also found that 0.3% of the patients studied were infected by two different genotypes, suggesting that carrier patients can be reinfected by HCV at a later time. Although some authors have argued that the frequency of mixed infections in serum with different HCV types is very low, even in high-risk groups,19 others have found simultaneous nosocomial spreading of two different strains in one hemo-dialysis unit,20 supporting the argument of simultaneous infection with at least two HCV types. Recently, Idrees and Riazuddin found a frequency of mixed HCV infection of 4.8% in patients studied in Pakistan.21
The wide regional classification throughout Mexico used in our study provides information about the variations in HCV genotypes throughout the country. First, we found a trend in the distribution of genotype 1 frequencies from the high rates in the North-East, North, and Center-East regions to the lowest frequencies in the South, East, and Peninsula regions. The northern part of the country did not have a homogeneous frequency of genotype 1 but instead showed frequencies ranging from 81.2% in the North-East, 73.6% in the North, and 69.4% in the North-West. Second, genotype 2 occurred with higher frequencies in the Peninsula, East, and South regions than in the North-West, North, and North-East regions, which had the lowest frequencies. Third, the prevalence rates of genotype 2 were fairly similar in the North-East, North, and NorthWest regions (12.3%, 14.1%, and 15.9%, respectively). Fourth, although genotype 3 accounted for only 7.2% of all the cases studied, we found significant differences in the prevalence distribution between regions. The North-West and North regions had the highest prevalence rates of genotype 3 (13.8% and 11.5%, respectively), and the probability of infection by genotype 3 was 20 times (95% CI 2.5, 125) and 10 times (95% CI 3.8, 25) higher in these regions than in the South region, which had a prevalence of 0.9%. With genotype 3 being strongly associated with intravenous drug usage, this might suggest that the main source of infection could be related to it.
Our results have important implications for therapeutic intervention: genotype 1 is the most prevalent in Mexico, accounting for 70.3% of cases on average. Because genotype 1 is one of the most resistant types to pegylated interferon and ribavirin treatment,22 early diagnosis and therapy are important to attain high rates of sustained SVR.23 Early diagnosis implies the identification of low HCV RNA levels. According to Jensen, et al,6 patients with a baseline HCV RNA of < 200,000 IU/mL (low level) have a 2.7-times higher probability of an SVR (95% CI 1.1, 6.3) than patients with a viral load > 600,000 IU/mL (high level). Patients with genotype 1 and a low HCV RNA load have almost an 89% probability of an SVR after 24 weeks of treatment.6 We found that only 22.4% of patients with genotype 1 in Mexico were classified in the low-load category after HCV RNA quantification. Among these patients, women had a higher frequency (24.9%) than men (19.7%) (p = 0.014). There were few differences between regions in the frequency rates of patients with HCV genotype 1 in the low-load category of HCV RNA. These results suggest that prevention and health promotion programs to control chronic hepatitis C in Mexico could be improved by increasing early diagnosis and treatment, especially among patients with genotype 1 and among men, independently of the region in which they were diagnosed.
These findings could also help develop drug-utilization strategies to improve the economic burden of treating chronic hepatitis C. Genotype 2 can be treated successfully with conventional interferon and could be used more widely in the South, East, and Peninsula regions.24
Finally, the results of our study show that more than one HCV genotype exists in Mexico and that each of these genotypes has few subtypes (three for genotype 1, four for genotype 2, and three for genotype 3). The first three genotypes occur most frequently in Mexico and account for 99.3% of all patients studied. Thus, these genotype distributions show a reduced diversity of subtypes, suggesting that they have been introduced relatively recently to Mexico and that their geographic distribution is related to the pattern of introduction.
Abbreviations- •
HCV. Hepatitis C Virus.
- •
RNA. Ribonucleic Acid.
- •
RT-PCR. Reverse transcription polymerase chain reaction.
- •
OR. Odds Ratio.
- •
CI. Confidence Interval.
- •
IU/mL. International Units per Mililiter.
This trial was partially supported by a Grant from Grupo Roche Syntex de Mexico. Ricardo Jimenez-Mendez was an employee of Grupo Roche Syntex de Mexico at the time this study was performed.