Hepatocellular carcinoma (HCC) is one of the most common and fatal tumors in the world, ranking third in cancer-related mortality. Chronic HBV infection is one of the major risk factors for hepatocellular carcinoma in China, Korea, and Sub-Saharan Africa. The HBx protein encoded by the X gene of HBV is a broadly regulated protein involved in transcriptional activation, epigenetics, apoptosis, DNA repair, and other regulatory processes. This study aimed to investigate the mechanism of HBx regulation of miR-155 and PTEN (Phosphatase and tensin homolog deleted on chromosome ten) in HBV-HCC.
MethodsExosomal miR-155 quantity was analyzed by sampling serum exosomes of patients with hepatocellular carcinoma and normal subjects. The analysis was divided into different subgroups according to HBV positivity or negativity. At the cellular level, the biological roles of HBX, microRNA-155 and PTEN on hepatocellular carcinoma cells and their regulatory relationships with each other were verified.
ResultsMicroRNA-155 and PTEN expression in HBV-positive HCC liver cancer tissues were negatively correlated, and HBX and miR-155 expression were positively correlated; microRNA-155 could target and inhibit PTEN expression, thereby promoting hepatocellular carcinoma cell activity, inhibiting apoptosis, and promoting invasion and migration; HBX could upregulate microRNA-155 thereby inhibit PTEN to promote malignant transformation of hepatocellular carcinoma.
ConclusionsHBX could promote malignant transformation of hepatocellular carcinoma cells by upregulating microRNA-155 expression and thereby inhibiting the PTEN/PI3K-AKT pathway. Blocking miR-155 expression could attenuate the proliferation-promoting and invasive effects of HBX.
Hepatocellular carcinoma (HCC) is one of the most common fatal tumors in the world, ranking third in cancer-related mortality and sixth in major cancer types [1]. In China, Korea and Sub-Saharan Africa, the major risk factors for liver cancer are chronic HBV infection and aflatoxin exposure or both [1]. HBV is a semi-double-stranded circular DNA virus with four overlapping open reading frames (ORFs) in its genome encoding protein S (viral surface protein), protein C (viral core protein), protein P (viral polymerase) and protein X (HBx protein). The X gene encodes the HBx protein, a protein with a wide range of regulatory roles involved in transcriptional activation, epigenetics, apoptosis, DNA repair, and other regulatory processes [2]. For example, Song et al [3]. showed that the mammalian Zinc fingers and homeoboxes 2 (ZHX2) gene was significantly reduced in tumor tissues of HBV-positive hepatocellular carcinoma patients and in the livers of HBV-transgenic mice and that HBV inhibited ZHX2 expression to promote HCC proliferation. HBx-mediated upregulation of TRERNA1 promotes sorafenib resistance and cell proliferation in hepatocellular carcinoma by sponge action miR-22-3p targeting NRAS [4].
Exosomes are small extracellular membrane vesicles(EVs) containing nucleic acids and proteins excreted by HSCs and mesenchymal stem cells [5,6]. EVs enter the intercellular space or enter body fluids such as urine, saliva, or blood, which may be taken up by neighboring cells or potentially delivered to target cells [7]. Exosomes are a readily accessible and rich source of biomarkers that can assess organ disease, including that of the liver [8]
Small molecular ribonucleic acids (microRNAs) are small non-coding RNAs of –18 to 25 nucleotides. miR-155 is located within exon 3 of the B cell integration cluster (BIC), a non-coding gene on human chromosome 21, and consists of a short-stranded non-coding RNA of 23 nucleotides [9]. miR-155 is a multifunctional microRNA, and aberrant miR-155 expression affects the development of inflammatory and autoimmune diseases and plays an important role in tumor cell proliferation, differentiation, migration, apoptosis, and the body's tumor immune response. Our previous findings showed that exosomal miR-155 was closely associated with the progression of cirrhosis and clinical prognostic indicators of cirrhosis and gradually increased with the severity of liver necrosis and fibrosis [10]. In addition, a study by Fu et al [11]. showed that microRNA-155-5p inhibited PTEN through the PI3K/Akt pathway to promote hepatocellular carcinoma progression. However, little is known about the involvement of miR-155 in regulating the development of HBV-HCC, and the regulatory mechanism between HBX and miR-155 is unclear.
This study aimed to investigate the mechanism of HBx regulation of miR-155 and PTEN in HBV-HCC. our study found that miR-155 inhibited PTEN to promote malignant transformation of hepatocellular carcinoma and could serve as a bridge for HBX regulation of PTEN.
2Material and methods2.1Human liver cancer tissue samples and serum samplesA total of 190 patients with HCC treated at the Department of Hepatobiliary Surgery, Tianjin First Central Hospital from October 2013 to August 2015 were collected in this study, along with peripheral blood, cancerous and paracancerous tissues, including HBV-positive patients in 102 cases, HBV-negative patients in 79 cases and HCV-positive patients in 9 cases (not included in the study). Peripheral plasma was collected from each HBV-positive and HBV-negative patient and transferred to cuvettes. Tubes were centrifuged at 1,200 × g for 15 min at 26°C. After plasma centrifugation, samples were stored in liquid nitrogen until use, and cancer and paracancerous tissues were similarly stored in liquid nitrogen. All procedures involving human participants in this study were in accordance with the Declaration of Helsinki (revised 2013). The Institutional Review Board of Tianjin First Central Hospital authorized the study protocol. All participants provided written informed consent prior to recruitment, and the follow-up deadline was January 2021.
2.2Exosome extraction and identificationExosome extraction and identification,Transmission electron microscopy (TEM) to verify exosome morphology were performed as previously described [10].
2.3Quantitative reverse transcription polymerase chain reaction (qRT-PCR)Total RNA was isolated from cancerous tissues, paracancerous tissues and exosomes. To validate miRNA or mRNA expression, qRT-PCR was performed using a SYBR Premix Dimereraser kit (TaKaRa Biotechnology, Kyushu, Japan) on a LightCycler 480 II detection system (Roche Diagnostics, New Naxi, USA). The primers for miR-155,PTEN,HBX and GAPDH were purchased from Sangon Biotech (Shanghai, China). The primers are listed in Table 1. The relative gene expression values for the target miRNA were calculated using the 2–ΔΔCT method.
Primer sequences used for quantitative reverse transcription polymerase chain reaction.
Cells were cultured in antibiotic-free growth medium in 6-well plates. According to the manufacturer's instructions after 24 hours of incubation, the cells were transfected using Lipofectamine 2000. the growth medium was changed after 6 hours. the transfected cells were harvested after 48 hours for the next step of the experiment.
2.5Construction of sTable transfer cell linesCells in good condition were spread into six-well culture plates and incubated at 37°C, 5% CO2 for 12h; the supernatant was removed and incubated overnight at 37°C, 5% CO2 according to 2ml of fresh medium without P/S and 3μl polybrene (8mg/ml) 1ml virus solution per well; after 48h of virus infection, the corresponding antibiotics were added to each well to kill the uninfected cells and continuously maintained for several days until all cells were killed using the wild-type cell line as a control.
2.6Western blotTreated cells were lysed in RIPA buffer (Heart, Beijing, China). Total protein samples were separated by SDS-PAGE polyacrylamide gels and transferred to PVDF membranes (Millipore, Billerica, MA, USA). Membranes were immunized overnight at 4°C with the following primary antibodies, PTEN, HBX, cyclinD, E-cadherin, E-cadherin (WanleiBio, Shenyang, Liaoning, China); PVDF membranes were washed with TBST and then incubated with secondary antibody, HRP-conjugated goat IgG (1:5000; Transgene, China) at 37°C for 1 hr. Signals were detected with a Bio-Rad gel imaging system. Images were quantified by Quantity One (Bio-Rad, USA) and the relative protein expression in each sample was normalized to β-actin levels.
2.7CCK8 assayCells were added to a 96-well plate, five assay time points were set, 0, 12, 24, 48 and 72h, three replicate wells were set for each group, placed at 37°C, 5% CO2 and incubated at a constant temperature for 12h, then the plasmid was transfected into the cells. The growth medium was changed 6 h after transfection and the cells continued to be cultured for 48 h. The supernatant was removed and added to the six-well plate according to the dose of 200 ul fresh medium and 10 μl CCK8 reagent and incubated under the same conditions as above for 2 h. The absorbance values at OD of 450 nm were detected with an enzyme marker, and the cells were tested at 12 h, 24 h, 48 h and 72 h, respectively after After the same incubation as described above, the absorbance values at an OD value of 450 nm were detected.
2.8Apoptosis assayThe concentrated cells were stained with PE Annexin V Apoptosis Assay Kit (BD Biosciences, USA), washed twice with PBS, resuspended by adding 100ul of binding buffer, added 5ul of PE Annexin V and 5ul of 7-AAD to the cell suspension, and stained for 15 min with protection from light. 400ul of binding buffer was added at the end of staining, and flow cytometry (BD influx Cell Sorter,BD Biosciences, San Jose, CA, USA) assay was completed within 1 hour and analyzed by FACS software.
2.9Wound-healingCells were seeded in 6-well plates. At 48 h post-transfection, cell layers in serum-free medium were scratched using a 10ul tip. After 24h incubation, three areas were randomly observed for migration comparison and the experiment was repeated three times. Scratch gap width before and after analysis using ImageJ software.
2.10transwell assaysTranswell membranes (Millipore, USA) coated with Matrigel and serum-free medium were used to detect cell invasion. At 24 h post-transfection, cells were cultured into serum-free medium and re-added to the upper layer, and 10% FBS medium was added to the lower layer as a chemoattractant. After 24 h of incubation, the bottom cells were stained with 0.1% crystal violet (Beyotime, Shanghai, China), and images of Transwell cell invasion were acquired with an inverted microscope.
3Statistical analysisAll data from this experiment were statistically analyzed using SPSS 24.0 (IBM, Armonk, NY, USA); mean ± standard deviation (x±s) was used to express the measurement data;t-test was used for comparison of means between two groups; correlation between variables was analyzed by linear correlation. p<0.05 was considered a statistically significant difference.
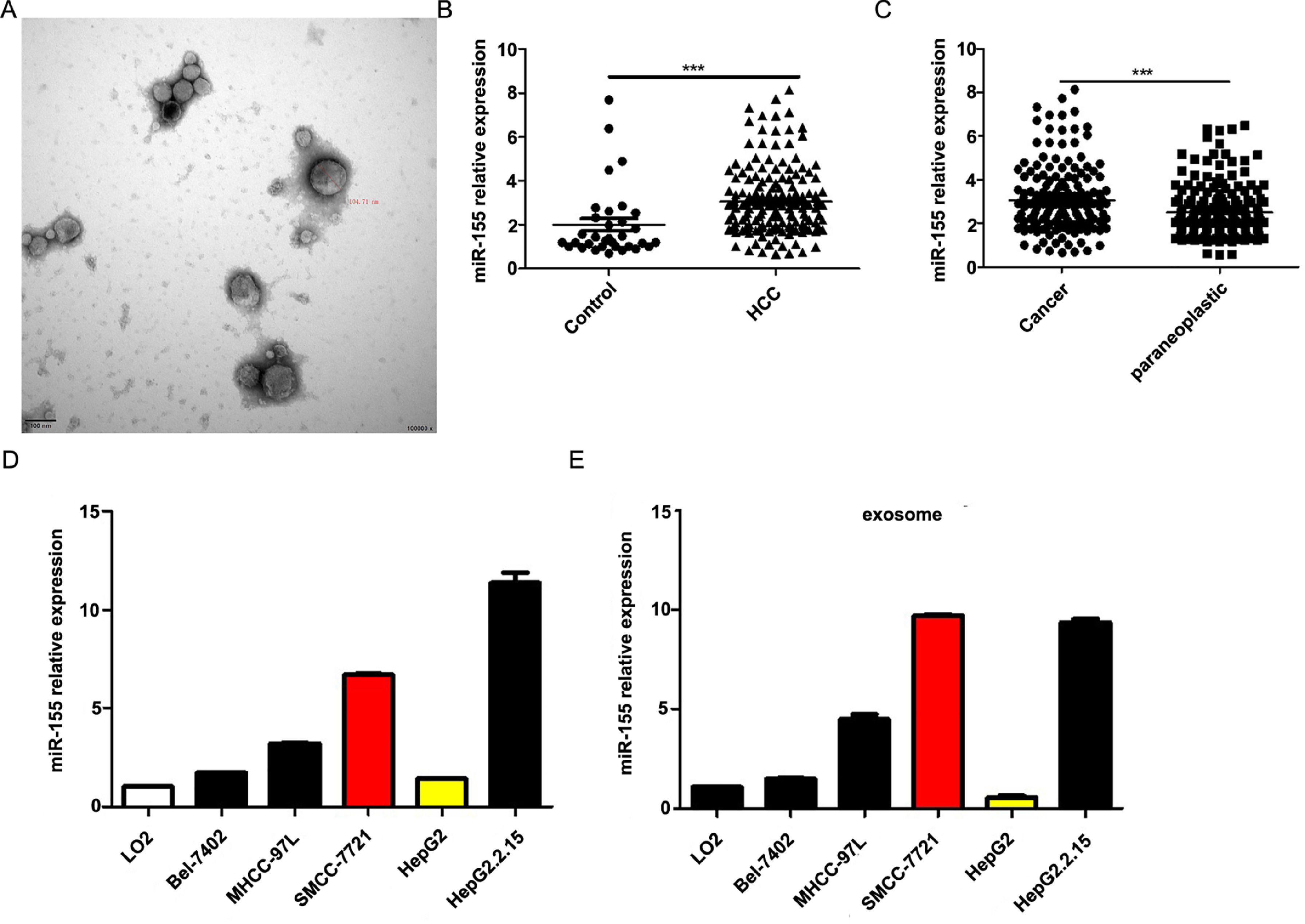
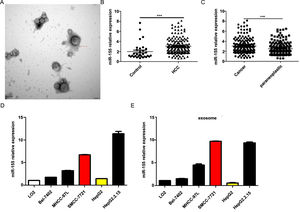
4Result4.1MiR-155 is highly expressed in HCC patient serum-derived exosomes, tumor tissues, and HCC cell linesOur previous study showed that exosomal miR-155 was closely associated with the progression of cirrhosis and clinical prognostic indicators of cirrhosis and gradually increased with the severity of liver necrosis and fibrosis. To further investigate the role of miR-155 in HCC disease progression, we examined miR-155 in 33 normal, 79 HBV- and 102 HBV+ HCC patients with serum exosomes, and we also examined the relative expression levels of miR-155, HBX, and PTEN in these 102 HBV-positive HCC cancer tissues and paracancerous tissues. The results showed that electron microscopy confirmed that the diameter of extracted exosomes ranged from 80-120 nm (Fig. 1A), and the expression of plasma exosomal miR-155 was significantly increased in HCC patients compared with controls (P < 0.001, Fig. 1B), and the relative expression of miR-155 was significantly higher in HCC cancer tissues than in paraneoplastic tissues (P < 0.001, Fig. 1C). Next, we evaluated miR-155 expression in five HCC cell lines (including HepG2, Bel-7402, MHCC-97L, SMMC-7721, and HepG2.2.15) and an immortalized hepatocyte line L-02, and the results showed that miR-155 expression was relatively low in HepG2 cells, while in SMMC-7721 cells with relatively high expression levels, and miR-155 expression was highest in the HBV-positive HepG2.2.15 cell line, with similar conclusions drawn for the expression of cellular exosomal miR-155 (Fig. 1D).
MiR-155 Is Highly Expressed in HCC Patient Serum-Derived Exosomes, Tumor Tissues, and HCC Cell lines. A Plasma exosome electron microscopy images; B Differences in relative expression of plasma exosome miR-155 in HCC compared with controls; C Differences in relative expression of miR-155 in cancer and paraneoplastic tissues; D Relative expression of microRNA-155 within each cell line and in exosomes.
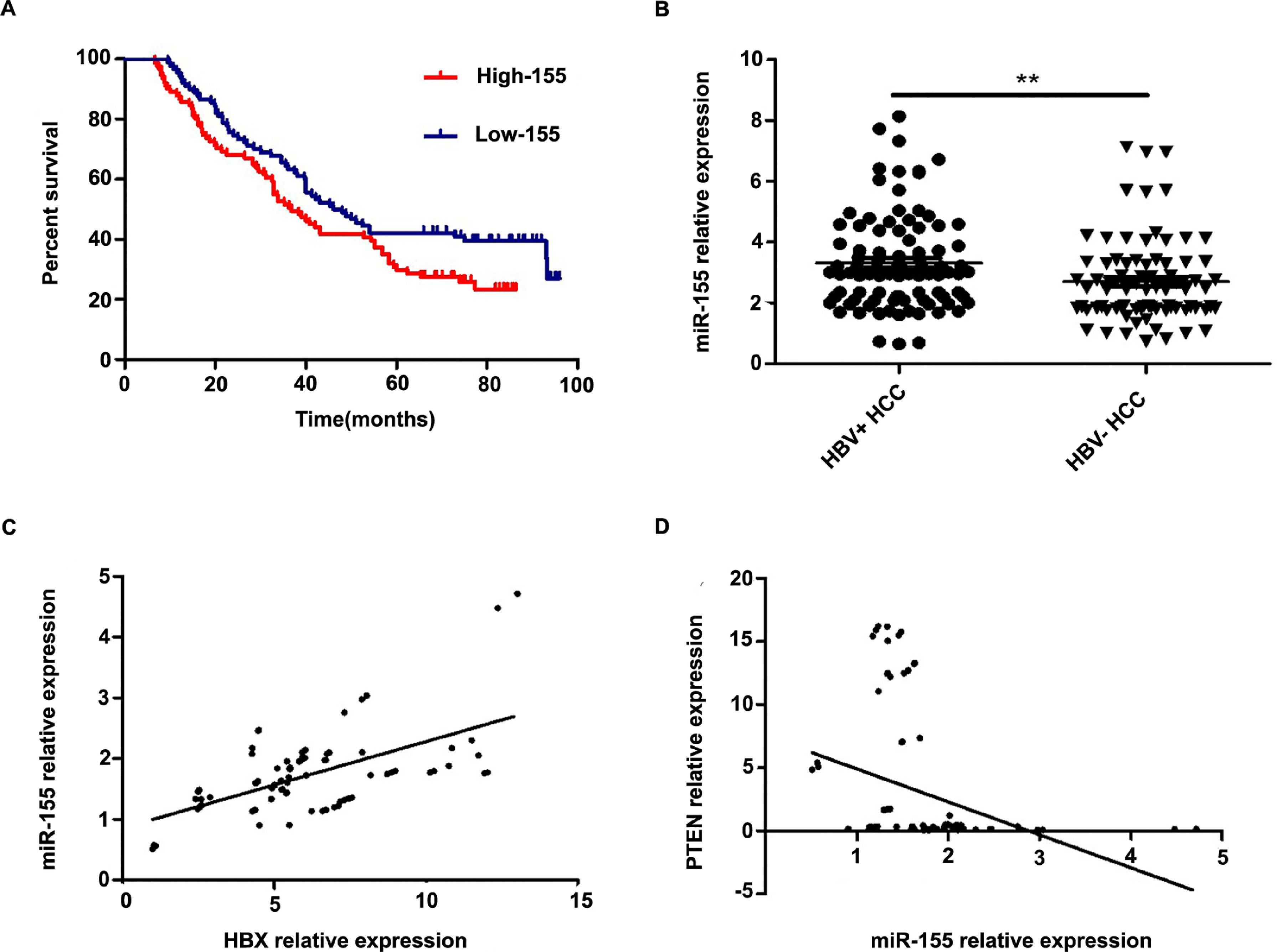
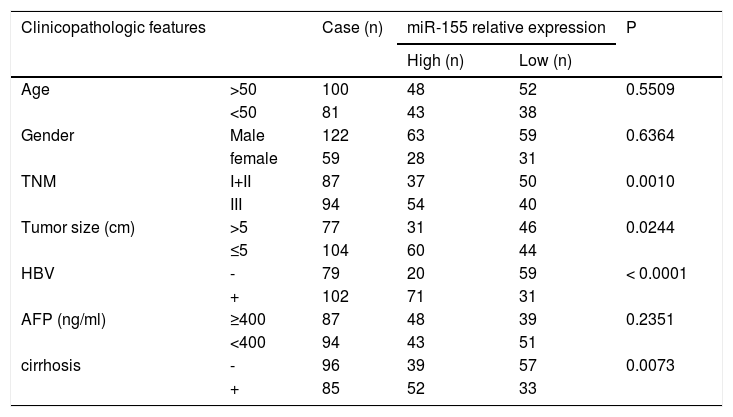
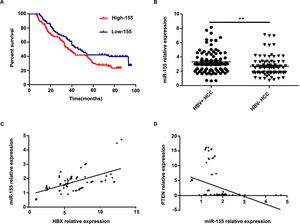
Further analysis combining miR-155 and clinical data revealed that the expression of exosomal miR-155 was closely correlated with tumor size, tumor stage, cirrhosis, and HBV positivity, while there was no statistical difference with age, AFP, and gender (Table 2). We divided HCC patients into two groups according to the median expression of plasma exosomal miR-155. The group with low expression of exosomal miR-155 had a longer survival time than the group with high expression (Fig. 2A). Plasma exosomal miR-155 expression was significantly higher in HBV+ HCC patients than in HBV-HCC patients (P=0.0048, Fig. 2B). Linear correlation analysis showed a significant positive relationship between HBX and miR-155 in cancer tissues (r=0.5788, P<0.001, 2C) and a significant negative relationship between miR-155 and PTEN (r=-0.3057, P=0.0018, 2 D). In summary, miR-155 was highly expressed in HBV-associated HCC plasma exosomes, cancer tissues and cancer cells and was positively correlated with HBx expression.
Relationship between exosomal microRNA-155 expression and clinicopathology in patients with hepatocellular carcinoma.
Exosomal miR-155 can be used as a marker of patient prognosis, and miR-155 expression in liver cancer tissues was closely correlated with HBX and PTEN. A high microRNA-155 (miR-155) expression group prognosis compared with the low-miR-155 expression group; B Differences in relative expression of HBV miR-155 in HBV+ and HBV-HCC patients versus controls; C Linear regression model of HBX and microRNA-155 in cancer tissue; D Linear regression model of microRNA-155 and PTEN in cancer tissue.
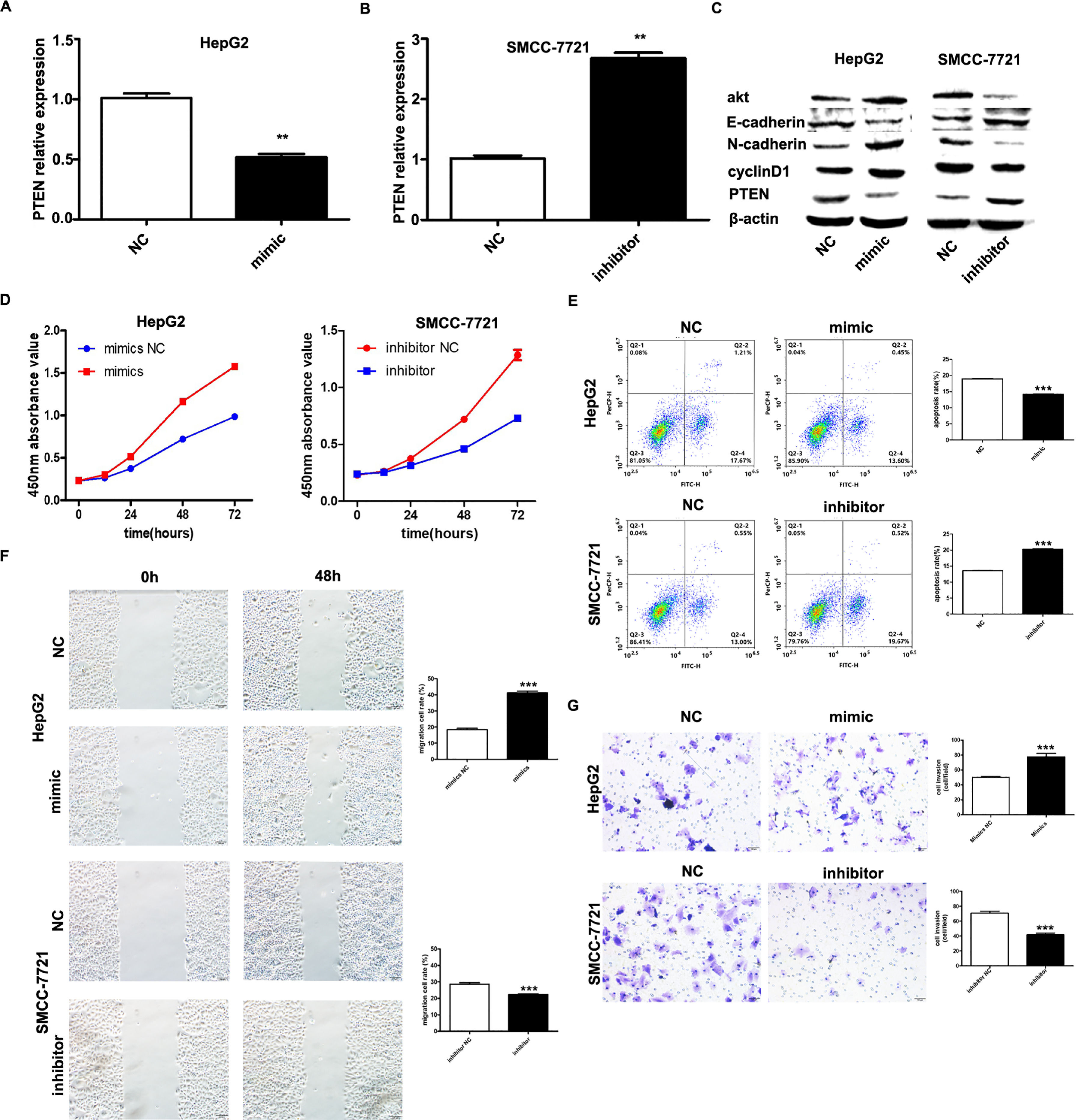
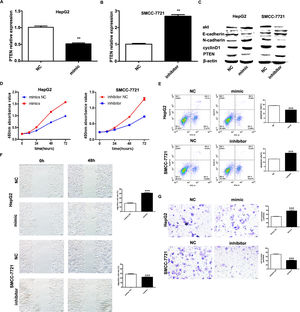
It has been shown that microRNA-155 can inhibit PTEN expression from promoting the proliferation of hepatocellular carcinoma cells and that PTEN is a target of miR-155; based on the expression of miR-155 in hepatocellular carcinoma cell lines, HepG2 and SMMC-7721 were selected as the cell lines for study. we investigated by transfecting miR-155 mimics into HepG2 cells and miR-155 inhibitor transfected into SMMC-7721 cells. qRT-PCR results showed that PTEN gene expression was down-regulated in HepG2 cells and upregulated in SMMC-7721 cells (Fig. 3A, B); western blot results showed that in the HepG2 mimic group PTEN, E- cadherin expression was down-regulated and cyclin D, N-cadherin expression was upregulated in the HepG2 mimic group, while the opposite result was obtained in the SMMC-7721 group (Fig. 3C). Further CCK8 assay showed that miR-155 high expression promoted the viability of HepG2 cells and miR-155 low expression inhibited the viability of SMMC-7721 cells (3D); flow assay results showed that miR-155 high expression inhibited the apoptosis of HepG2 cells, while miR-155 low expression promoted the apoptosis of SMMC-7721 cells (3E); furthermore, according to wound scratch assay and transwell assay, high expression of miR-155 resulted in significantly enhanced cell migration and invasion of HepG2 cells, with the opposite results observed in SMMC-7721 cells (Fig. 3F and G). In summary, miR-155 down-regulates and partially down-regulates PTEN expression promoting proliferation, migration and invasion of hepatocellular carcinoma cells and inhibiting apoptosis.
microRNA-155 inhibits PTEN to promote hepatocellular carcinoma cell biology. A qRT-PCR to detect the relative expression of microRNA-155 and PTEN in each group; B western blot to detect the expression of PTEN, E-cadherin, cyclin D, N-cadherin in each group; C CCK8 assay to respond to the C CCK8 assay; D Flow cytometry assay to detect apoptosis; E Scratch assay to detect migration ability; F traswell chamber assay to detect invasion level. (Subgroups: HepG2 NC, HepG2 mimics, SMMC-7721 NC, SMMC-7721 inhibitor).
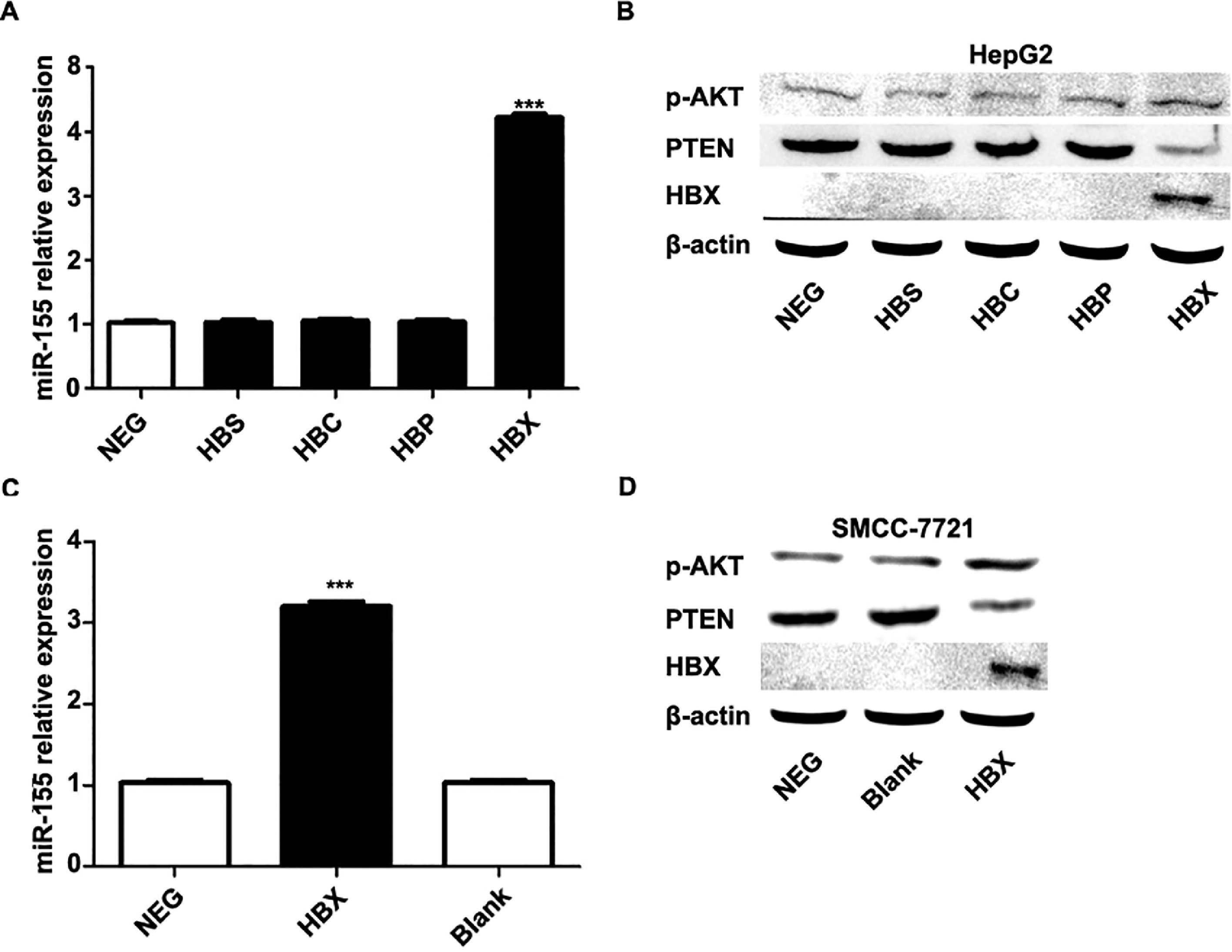
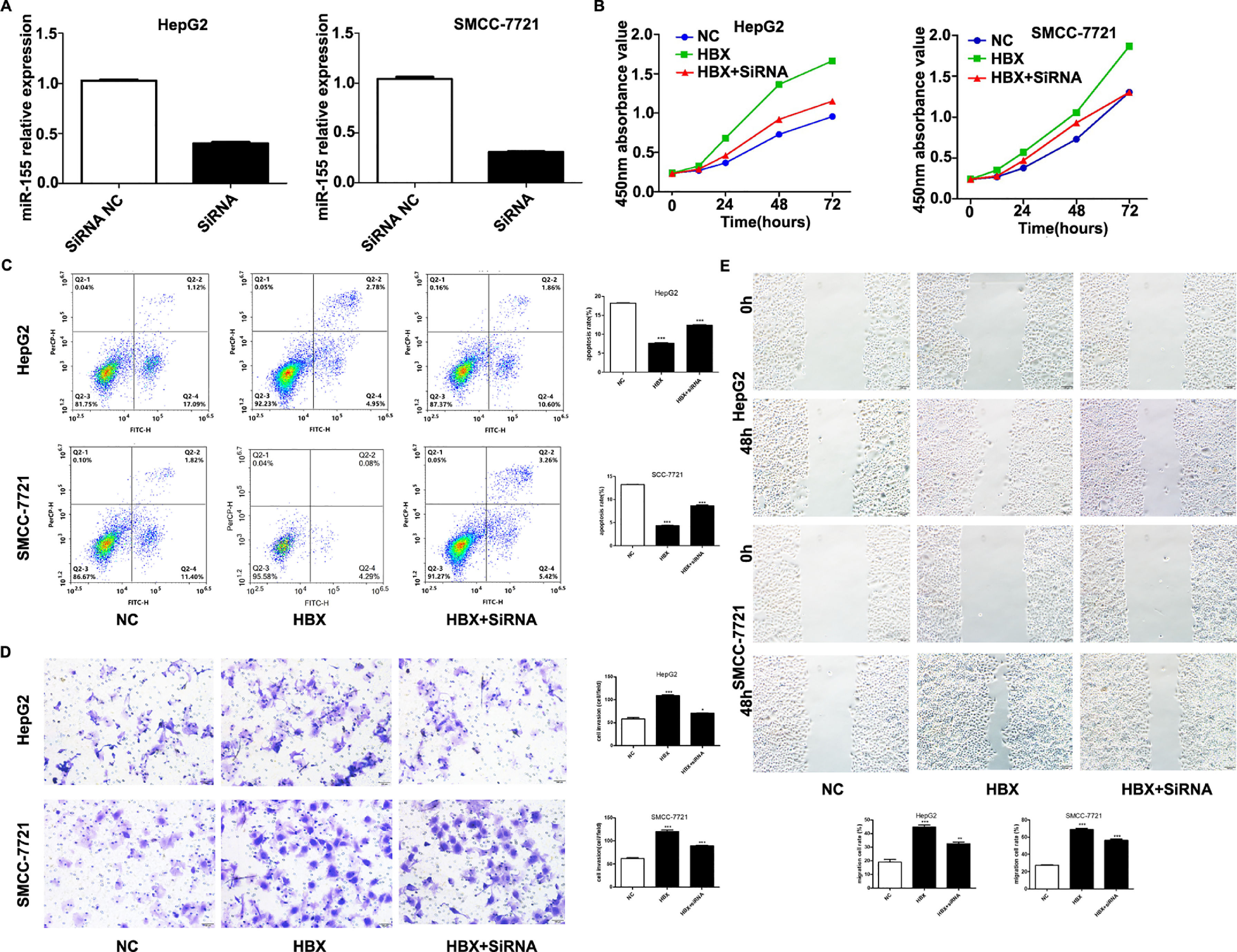
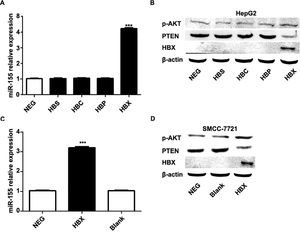
The previous experiments verified that miR-155 expression and HBV are closely related. In order to screen the HBV genes (HBS, HBP, HBX, HBC) that play a major role in promoting cancer, we investigated by constructing HepG2 cells stably expressing HBS, HBP, HBX, and HBC genes. qRT-PCR results showed that only high expression of HBX HepG2 cell lines showed elevated miR-155 expression, while the expression of miR-155 in cells high in HBS, HBP, and HBC showed no significant change compared to the control (Fig. 4A). Western blot results showed that high expression of HBX could inhibit PTEN expression and promote p-AKT expression (Fig. 4B). Similar results were obtained in the SMMC-7721 cell line by constructing a sTable transfer of HBX (Fig. 4C, D). CCK8 results showed enhanced viability of HepG2, SMMC-7721 cell line with high expression of HBX (5B); flow assay results showed that apoptosis was attenuated in HepG2, SMMC-7721 cell line with high expression of HBX (5C); transwell assay and scratch assay results indicated that HBX promoted migration and invasion of HepG2, SMMC-7721 cells (Fig. 5D, E).
HBX upregulates miR-155 to inhibit PTEN expression. A qRT-PCR to detect the relative expression of microRNA-155 and PTEN in HepG2 cells high in HBS, HBP, HBX, and HBC; B western blot to detect the relative expression of PTEN, p- AKT expression; C qRT-PCR to detect the relative expression of microRNA-155 and PTEN in SMMC-7721 cells with high expression of HBX; D western blot to detect the expression of PTEN, p-AKT in HepG2 cells with high expression of HBX.
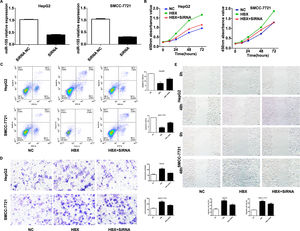
HBX upregulates miR-155 to inhibit PTEN expression A qRT-PCR to detect the relative expression of microRNA-155 and PTEN in HepG2 cells high in HBS, HBP, HBX, and HBC; B western blot to detect the relative expression of PTEN, p- AKT expression; C qRT-PCR to detect the relative expression of microRNA-155 and PTEN in SMMC-7721 cells with high expression of HBX; D western blot to detect the expression of PTEN, p-AKT in HepG2 cells with high expression of HBX (NC,HepG2-HBX,HepG2-HBX-shRNA; NC,SMMC-7721-HBX,SMMC-7721-HBX-shRNA).
We performed experiments by transfecting shRNA into HepG2, SMMC-7721 cell lines that stably overexpress HBX. qRT-PCR results showed decreased expression of miR-155 (Fig. 5A). CCK8 and flow assay showed that down-expression of miR-155 in HepG2, SMMC-7721 cell lines that overexpress HBX could attenuate cell viability and promote apoptosis (Fig. 5B, C), while low expression of miR-155 led to diminished migration and invasion ability of HepG2, SMMC-7721 cells with high expression of HBX according to transwell assay and scratch assay (Fig. 5D, E). Combined with the above findings suggest that HBX can promote the malignant transformation effect of hepatocellular carcinoma by upregulating or partially upregulating microRNA-155 and thus inhibiting the PTEN/p-AKT pathway.
5DiscussionPTEN is a negative regulator of intracellular phosphatidylinositol triphosphate levels, and one of the mechanisms by which PTEN inhibits cancer is dephosphorylation of PIP3, thereby inhibiting the PI3K/PKB/AKT signaling pathway to inhibit cell growth and promote apoptosis [12]. Decreased PTEN expression predicts progression and poor prognosis of HCC [13]. Our previous findings suggest that exosomal MiR-1290 promotes angiogenesis in hepatocellular carcinoma by targeting SMEK1 [14], and exosomal miR-155 is closely associated with the progression of cirrhosis and clinical prognostic indicators of cirrhosis and progressively increases with the severity of liver necrosis and fibrosis [10]. To further investigate the biological function of miR-155, we conducted a series of experiments to obtain a negative correlation between microRNA-155 and PTEN expression in HBV+ positive HCC liver cancer tissues. microRNA-155 could target and inhibit PTEN expression from promoting hepatocellular carcinoma cell activity, promoting invasion and migration, and inhibiting apoptosis. Numerous studies have also confirmed the biological role of miR-155. Zhang et al [15]. also showed that microRNA-155 could target ARID2 to promote human hepatocellular carcinoma proliferation. Su et al [9]. suggested that ectopic expression of miR-155 upregulated the expression of several IFN-induced antiviral genes in human hepatocellular carcinoma cells, and overexpression of miR-155 inhibited the expression of a repressor of cytokine signaling 1 (SOCS1) expression, which in turn enhanced the signal transducer and activator of transcription 1 (STAT1) and the signal transducer and activator of transcription 3 (STAT3) phosphorylation. they further demonstrated that miR-155 ectopic expression suppressed HBV X gene expression in vitro to some extent. microRNA-155 ectopic expression enhanced the natural antiviral immunity of human hepatoma cells against HBV infection.
Numerous studies have identified aberrant expression of relevant miRNAs in HBV-HCC. Aberrant expression of miRNAs plays an important biological role by regulating downstream target genes. HBx can play a key role in regulating the biological functions of hepatocellular carcinoma cells by regulating miRNAs or LncRNAs; HBx-mediated upregulation of TRERNA1 can act through sponge miR-22-3p targets NRAS to promote sorafenib resistance and cell proliferation in hepatocellular carcinoma [16]. HBx promotes HCC cell proliferation through downregulation of miR-145, miR-216b, miR-329 and miR-1236 [17–19]. Zhao et al [20]. reported that the oncoprotein astrocyte elevated gene-1 AEG-1 was overexpressed in various tumors, including hepatocellular carcinoma, and played an important role in promoting cell migration and invasion. HBx increased oncoprotein AEG-1 by downregulating miR-375 and miR-136 protein AEG-1 expression levels and promoting HCC cell migration. wang et al [21]. showed that HBX upregulated miR-155 expression in Kupffer cells (KCs) through PI3K and NF-kappaB signaling pathways. miR-155 increase promoted BCL-6, SHIP-1 and SOCS-1 expression by suppressing the production of HBx-induced inflammatory cytokines, thereby accelerating liver injury. The results of this study showed that microRNA-155 expression was closely associated with HBV positivity and positively correlated with HBX expression in cancer tissues. By establishing HepG2 and SMCC-7721 cell lines that stably overexpressed HBX and remediation experiments, we verified that HBX could upregulate microRNA-155 to promote hepatocellular's proliferation, invasion, and migration carcinoma cells.
In conclusion, this study reveals that HBX can promote malignant transformation of hepatocellular carcinoma cells by upregulating microRNA-155 expression and thereby inhibiting the PTEN/PI3K-AKT pathway at the clinical and cellular levels, and blocking miR-155 expression can attenuate the proliferation-promoting and invasive effects of HBX, which may provide a new therapeutic strategy for the treatment of HBV-associated hepatocellular carcinoma.
Ethical StatementThe authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The Institutional Review Board of Tianjin First Central Hospital authorized the study protocol. All participants provided written informed consent ahead of recruitment.
CRediT authorship contribution statementLian-Jie Niu: Conceptualization, Visualization, Funding acquisition, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. Tao Huang: Methodology, Writing – original draft, Writing – review & editing. Lianjiang Wang: Funding acquisition, Writing – original draft, Writing – review & editing. Xian-Fu Sun: Funding acquisition, Writing – original draft, Writing – review & editing. Ya-Min Zhang: Writing – original draft, Writing – review & editing.
The present study was supported by Henan Province Medical Science and Technology Tackling Program Project (NO: LHGJ20200159, LHGJ20200156) and the Science and Technology Fund of Tianjin Health Bureau(ZC20218).