Hepatotoxicity during tuberculosis (TB) treatment is frequent and may be related to the Arylamine N-Acetyltransferase (NAT2) acetylator profile, in which allele frequencies differ according to the population. The aim of this study was to investigate functional polymorphisms in NAT2 associated with the development of hepatotoxicity after initiating treatment for TB in people living with HIV/AIDS (PLWHA) in Pernambuco, Northeast Brazil.
Material and methodsThis was a prospective cohort study that investigated seven single nucleotide polymorphisms located in the NAT2 coding region in 173 PLWHA undergoing TB treatment. Hepatotoxicity was defined as elevated aminotransferase levels and identified as being three times higher than it was before initiating TB treatment, with associated symptoms of hepatitis. A further 80 healthy subjects, without HIV infection or TB were used as a control group. All individuals were genotyped by direct sequencing.
ResultsThe NAT2*13A and NAT2*6B variant alleles were significantly associated with the development of hepatotoxicity during TB treatment in PLWHA (p<0.05). Individual comparisons between the wild type and each variant genotype revealed that PLWHA with signatures NAT2*13A/NAT2*13A (OR 4.4; CI95% 1.1–18.8; p 0.037) and NAT2*13A/NAT2*6B (OR 4.4; CI95% 1.5–12.7; p 0.005) significantly increased the risk of hepatotoxicity.
ConclusionThis study suggests that NAT2*13A and NAT2*6B variant alleles are risk factors for developing hepatotoxicity, and PLWHA with genotypes NAT2*13A/NAT2*13A and NAT2*13A/NAT2*6B should be targeted for specific care to reduce the risk of hepatotoxicity during treatment for tuberculosis.
Tuberculosis (TB) remains a serious public health problem both in Brazil and throughout the world, and discovering seropositivity for the human immunodeficiency virus (HIV) during the diagnosis of TB is common. Data from the World Health Organization (WHO) estimates that of the 10.4 million people who developed TB worldwide in 2016, 1.1 million (11%) also presented with HIV infection [1]. In 2017, the Brazilian Northeastern state of Pernambuco presented the highest incidence rate of TB (46/100,000 pop.) within the region, together with a TB-HIV coinfection percentage of 11%. This percentage is higher than the overall national rate (9.2%) as well as that of the Northeast region (8.2%) [2].
The occurrence of hepatotoxicity during the treatment of TB in people living with HIV/AIDS (PLWHA), who experience difficulties inherent to a combination of the two diseases, may cause a negative effect on the therapeutic regimen and in most cases, lead to the discontinuation of treatment [3]. In Brazil, the incidence rate of hepatotoxicity following the use of drugs for TB treatment amongst those infected with HIV range between 31% and 44% [4,5].
Isoniazid is one of the major drugs used to treat TB, as recommended by the Brazilian Health Ministry, and is a recognized cause of liver toxicity [6]. The N-acetiltransferase2 enzyme (NAT2), encoded by the NAT2 gene, initiates the metabolism mechanism of isoniazid in the liver by biochemical acetylation, which produces acetylisoniazid and isonicotinic acid. Curiously, the acetylation rate of isoniazid varies and depends on the genetic component of each individual, which may be classified into rapid and slow acetylators [7].
There are reports in the literature describing various polymorphisms in genes that encode enzymes associated with hepatic metabolism, such as cytochrome P450 2E1 (CYP2E1), glutathione S-transferase (GST) and manganese superoxide dismutase (MnSOD, SOD2), and that are predisposed to hepatotoxicity induced by several substances [8].
In effect, NAT2 polymorphism explains the bimodal frequency distribution after administrating identical doses of isoniazid to different individuals, i.e. one group of rapid acetylators and another of slow acetylators [9].
Despite considerable evidence from within several populations demonstrating that the relationship between developing hepatotoxicity and the distribution of NAT2 encoding alleles differs according to ethnicity, there are few studies that address this issue in Brazil, and involve in their entirety, PLWHA and the diagnosis of active tuberculosis [10–13]. The aim of this study was to investigate NAT2 functional polymorphisms [NAT2*5D, NAT2*6B, NAT2*7A, NAT2*11A, NAT2*12A, NAT2*13A and NAT2*14A] associated with the development of hepatotoxicity after initiating treatment for TB in PLWHA in the state of Pernambuco, Northeast Brazil.
2Materials and methods2.1Design, site and study populationThis was a prospective cohort study with 173 PLWHA diagnosed with active tuberculosis, of both sexes, aged between 20 and 67 years, treated at the Hospital Correia Picanço, in a specialized service for PLWHA in the state of Pernambuco, Northeast Brazil. In addition, a further 80 healthy individuals without HIV infection or tuberculosis were used as a control group.
2.2Ethical aspectsThis study was approved by the Research Ethics Committee at the Universidade Federal de Pernambuco, Brazil, under protocol No. 254/05. All participants in the study signed an Informed Consent Form, as required by the statement on ethics contained in Resolution N°466/12 of the Brazilian National Health Council.
2.3Biological samplesPeripheral vein blood samples (10mL) were obtained from all studied patients at baseline and at the end of the first and second months on the follow-up visits to the health service, irrespective of whether or not they presented symptoms of hepatitis, to measure the aspartate aminotransferase (AST) and alanine aminotransferase (ALT) enzymes.
Healthy individuals, enrolled on the National Register of Bone Marrow Voluntary Donors, were randomly selected. This group was composed of consanguineously unrelated individuals, matched by geographic region, so as to demonstrate that NAT2 allele and genotype frequencies are present irrespective of HIV infection, tuberculosis or hepatotoxicity.
2.4Defining the study variablesHepatotoxicity was defined as elevated aminotransferase levels and identified as being three times higher than before initiating TB treatment, with associated symptoms of hepatitis. Symptoms were considered as the occurrence of jaundice, nausea, vomiting, dyspepsia and asthenia [14]. The reference values adopted were AST – 36UI/mL and ALT – 32UI/mL, according to the manufacturer's instructions (Targa 3000®).
The biological variables potentially related to hepatotoxicity were analyzed, such as: gender, age, skin color (self-reported) as a proxy variable for ethnicity and body mass index (BMI) categorized as malnourished (BMI<18.5kg/m2), eutrophic (18.5≤BMI≥24.9kg/m2) and overweight/obese (BMI≥25.0kg/m2).
The treatments for TB, according to the Brazilian Ministry of Health, were categorized as the RIP regimen (consisting of three drugs: rifampicin, isoniazid and pyrazinamide), and the newly recommended RIPE regimen (consisting of four drugs: rifampicin, isoniazid, pyrazinamide and ethambutol) and others [14].
The use of antiretroviral drugs at the time of initiating TB treatment was confirmed through medical records and categorized as: not using HAART, initiated HAART before TB treatment and initiated HAART during the first two months of TB treatment. For the analysis, we considered a CD4 count measured in the six months prior to initiating TB treatment, and the variable was categorized as <200cells/mm3 and ≥200cells/mm3.
Patients were considered HIV-seropositive when they tested positive in two sequential tests, in accordance with the recommendations of the Ministry of Health in Brazil. The results of all the 346 tests for HIV antibodies conducted on the 173 PLWHA patients included in this study were positive.
All patients who had initiated TB treatment due to clinical diagnosis or after a bacteriological test confirmation were said to have TB infection.
2.5Laboratory proceduresThe serological test used for the detection of HIV infection was the Rapid-check HIV1 & 2 (NDI-Núcleo de Doenças Infecciosas, Vitória, ES, Brasil) and the rapid test – HIV-1/2 Bio-Manguinhos (Fundação Oswaldo Cruz/Biomanguinhos, Rio de Janeiro, Brasil).
The CD4+T lymphocyte count was performed by flow cytometry using anti-CD4 antibodies labeled with fluorescent dyes through the BD FACSCalibur™ system.
2.6NAT2 polymorphism analysisNAT2 genotypes were defined by direct sequencing, comprising two loci. Genomic DNA isolation was performed using the Biopur Extraction Kit – Mini Cent (Biometrix®, Brazil) according to the manufacturer's specifications. After extraction, DNA samples were stored at −20°C until use.
PCRs were performed with two independent reactions, as follows: 36.5μl H2O, 5μl 10× PCR buffer, 2μl MgCl2 50mM, 1μl 10mM dNTP, 2μl of each 10mM primer, 1μl DNA, and also 0.5μl Taq (5U/μl) to 50μl final volume. The single nucleotide polymorphisms (SNPs) G191A (NAT2*14A), C282T (NAT2*13A) and T341C (NAT2*5D) were evaluated using the 5′-CCTTACAGGGTTCTGAGACTAC-3′ and 5′-GGTGCCTTGCATTTTCTGCT-3′ pair primers and the following cycling parameters: initial denaturation (94°C/3min), 35 cycles (94°C/1min, 50°C/1min, and 72°C/1min), and final extension at 72°C/5min. On the other hand, the SNPs C481T (NAT2*11A), G590A (NAT2*6B), A803G (NAT2*12A) and G857A (NAT2*7A) were evaluated using 5’-CATTGTCGATGCTGGGTCTG-3 and 5’-TCATCCATAAAAATGTCAGCATTT-3′ primers with the same cycling parameters listed above, except for the annealing temperature (55°C).
The 667bp and 665bp amplicons were amplified respectively and analyzed by electrophoresis in 1% agarose gel. Direct sequencing was performed using the ABI PRISM BigDye Terminator v.3.1 Sequencing Kit (PE Applied Biosystems). Genotyping and sequence analysis were performed using the ApE-A-plasmid-Editor v2.0.45 with the GenBank accession n° AY331807 sequence as a reference (<www.ncbi.nlm.nih.gov/nuccore/AY331807>), following the International Committee for Gene Nomenclature NAT2 (<http://nat.mbg.duth.gr>).
From amongst the 7 SNPs commonly found in the human population, for predicting genotypes and phenotypes, four SNPs, which result in amino acid substitution and the consequent decrease in the acetylation capacity, were analyzed. Individuals who presented at least two variant alleles in any of the SNPs at positions 191A (NAT2*14A), 341C (NAT2*5D), 590A (NAT2*6B) and 857A (NAT2*7A) were classified as slow acetylators [15].
The reference SNPs (rs) for the alleles NAT2 studied were, respectively: rs1801280 (NAT2*5D), rs1799930 (NAT2*6B), rs1799931 (NAT2*7A), rs1799929 (NAT2*11A), rs1208 (NAT2*12A), rs1041983 (NAT2*13A) and rs1801279 (NAT2*14A).
2.7Statistical analysisData were stored in a database created for this research. All data were double entered (validated in EPI-INFO 6.04), and were subsequently compared to identify any possible typing errors.
The analysis was performed using STATA 12.0 (Statistical Software for Professionals, StataCorp LP, UK). To check the statistical significance of differences in the frequency distribution of variables and the Hardy–Weinberg equilibrium, the chi-square test was used, or when necessary, the Fisher exact test. The bivariate logistic regression analysis was performed and the magnitude of the associations was expressed by the odds ratio (OR) as an estimate of relative risk, with a confidence interval of 95% regression and a significance level of 5% (p<0.05). Estimation for haplotypes frequency was based on the maximum likelihood method using Arlequin 3.5.
3ResultsAmongst the 173 patients enrolled in this study, 53 (30.6%) presented symptoms consistent with hepatitis and high aminotransferase levels, three times higher than those recorded before initiating TB treatment. These were the cases of hepatotoxicity.
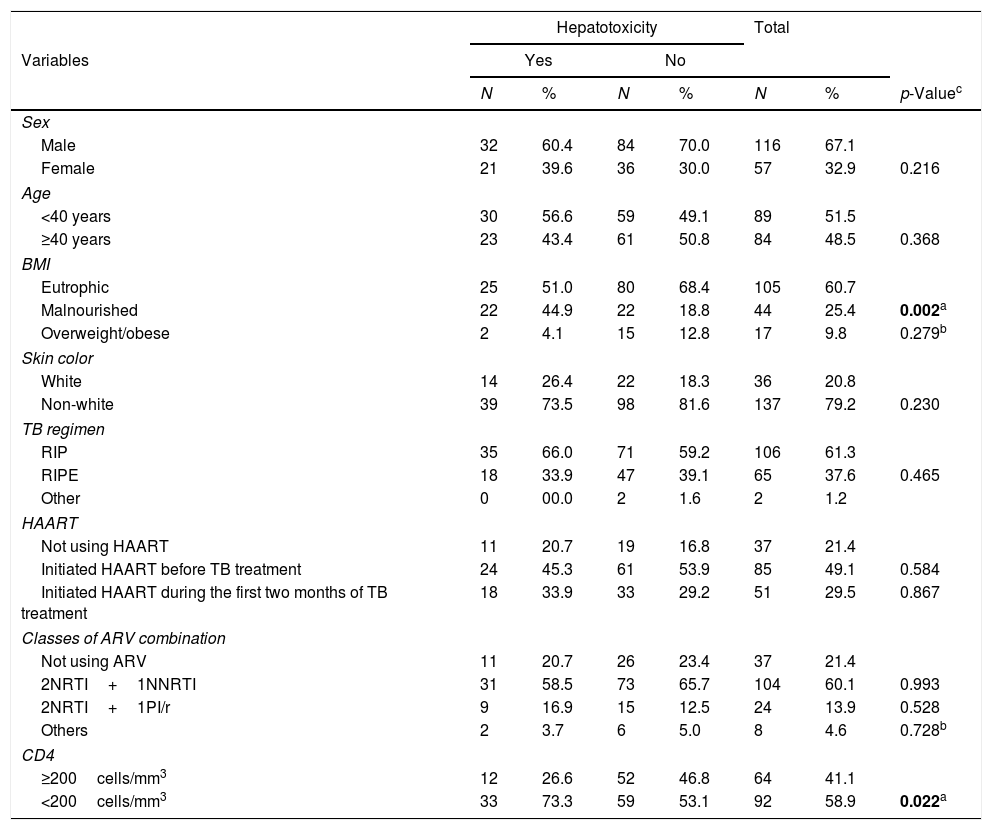
Table 1 demonstrates the frequency distribution of biological and clinical variables of PLWHA with tuberculosis. This cohort was predominantly male (67%), just over half (52%) were aged under 40, with a greater proportion of eutrophic (61%) and non-white (79%) individuals. It was found that the most commonly used therapeutic regimen for TB (61%) was that of three drugs (i.e., RIP – Rifampicin, Isoniazid and Pyrazinamide), and HAART (Highly active antiretroviral therapy) was being used by 78.6%, the majority of whom had initiated HAART before treatment for TB (49%). The antiretroviral regimen containing two NRTIs (nucleoside reverse transcriptase inhibitors) and one NNRTI (non-nucleoside reverse transcriptase inhibitor) was the most commonly used by HAART users (60%). Slightly more than half (59%) the population presented a CD4 level of less than 200cells/mm3 and this was statistically significant (p=0.022) (Table 1).
Clinical and biological variables of PLWHA with tuberculosis. Recife, Pernambuco, Brazil.
| Hepatotoxicity | Total | ||||||
|---|---|---|---|---|---|---|---|
| Variables | Yes | No | |||||
| N | % | N | % | N | % | p-Valuec | |
| Sex | |||||||
| Male | 32 | 60.4 | 84 | 70.0 | 116 | 67.1 | |
| Female | 21 | 39.6 | 36 | 30.0 | 57 | 32.9 | 0.216 |
| Age | |||||||
| <40 years | 30 | 56.6 | 59 | 49.1 | 89 | 51.5 | |
| ≥40 years | 23 | 43.4 | 61 | 50.8 | 84 | 48.5 | 0.368 |
| BMI | |||||||
| Eutrophic | 25 | 51.0 | 80 | 68.4 | 105 | 60.7 | |
| Malnourished | 22 | 44.9 | 22 | 18.8 | 44 | 25.4 | 0.002a |
| Overweight/obese | 2 | 4.1 | 15 | 12.8 | 17 | 9.8 | 0.279b |
| Skin color | |||||||
| White | 14 | 26.4 | 22 | 18.3 | 36 | 20.8 | |
| Non-white | 39 | 73.5 | 98 | 81.6 | 137 | 79.2 | 0.230 |
| TB regimen | |||||||
| RIP | 35 | 66.0 | 71 | 59.2 | 106 | 61.3 | |
| RIPE | 18 | 33.9 | 47 | 39.1 | 65 | 37.6 | 0.465 |
| Other | 0 | 00.0 | 2 | 1.6 | 2 | 1.2 | |
| HAART | |||||||
| Not using HAART | 11 | 20.7 | 19 | 16.8 | 37 | 21.4 | |
| Initiated HAART before TB treatment | 24 | 45.3 | 61 | 53.9 | 85 | 49.1 | 0.584 |
| Initiated HAART during the first two months of TB treatment | 18 | 33.9 | 33 | 29.2 | 51 | 29.5 | 0.867 |
| Classes of ARV combination | |||||||
| Not using ARV | 11 | 20.7 | 26 | 23.4 | 37 | 21.4 | |
| 2NRTI+1NNRTI | 31 | 58.5 | 73 | 65.7 | 104 | 60.1 | 0.993 |
| 2NRTI+1PI/r | 9 | 16.9 | 15 | 12.5 | 24 | 13.9 | 0.528 |
| Others | 2 | 3.7 | 6 | 5.0 | 8 | 4.6 | 0.728b |
| CD4 | |||||||
| ≥200cells/mm3 | 12 | 26.6 | 52 | 46.8 | 64 | 41.1 | |
| <200cells/mm3 | 33 | 73.3 | 59 | 53.1 | 92 | 58.9 | 0.022a |
PLWHA (People living with HIV/AIDS), BMI (Body mass index), NAT2 (Arylamine N-Acetyltransferase), TB (Tuberculosis), RIP (Rifampicin, Isoniazid, Pyrazinamide), RIPE (Rifampicin, Isoniazid, Pyrazinamide, ethambutol), HAART (Highly active antiretroviral therapy), ARV (Antiretroviral therapy), NRTI (nucleoside reverse transcriptase inhibitor), NNRTI (Non-nucleoside reverse transcriptase inhibitor), PI/r (Protease inhibitor/ritonavir).
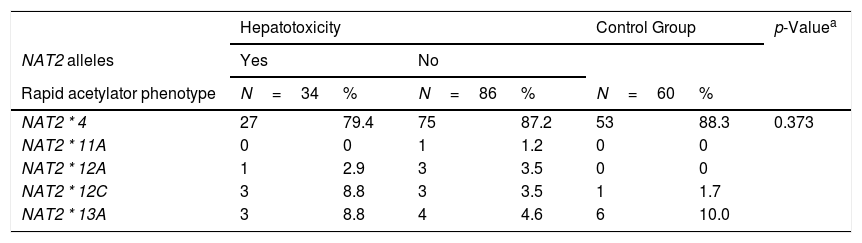
Seven different SNPs, previously known to be determinants of the acetylator phenotype were identified using NAT2 sequencing in the study population and control group (healthy individuals).
Table 2 describes the frequencies of these seven main mutations and their respective alleles related to the NAT2 acetylator phenotype profile.
Frequencies of NAT2 alleles in PLWHA with tuberculosis and control group. Recife, Pernambuco, Brazil.
| Hepatotoxicity | Control Group | p-Valuea | |||||
|---|---|---|---|---|---|---|---|
| NAT2 alleles | Yes | No | |||||
| Rapid acetylator phenotype | N=34 | % | N=86 | % | N=60 | % | |
| NAT2 * 4 | 27 | 79.4 | 75 | 87.2 | 53 | 88.3 | 0.373 |
| NAT2 * 11A | 0 | 0 | 1 | 1.2 | 0 | 0 | |
| NAT2 * 12A | 1 | 2.9 | 3 | 3.5 | 0 | 0 | |
| NAT2 * 12C | 3 | 8.8 | 3 | 3.5 | 1 | 1.7 | |
| NAT2 * 13A | 3 | 8.8 | 4 | 4.6 | 6 | 10.0 | |
| Slow acetylator phenotype | N=19 | % | N=34 | % | N=20 | % | |
|---|---|---|---|---|---|---|---|
| NAT2 * 5A | 1 | 5.3 | 1 | 2.9 | 1 | 5.0 | 0.976 |
| NAT2 * 5B | 7 | 36.8 | 14 | 41.2 | 7 | 35.0 | |
| NAT2 * 5C | 1 | 5.3 | 3 | 8.8 | 1 | 5.0 | |
| NAT2 * 5D | 5 | 26.3 | 9 | 26.5 | 4 | 20.0 | |
| NAT2 * 5J | 0 | 0 | 1 | 2.9 | 0 | 0 | |
| NAT2 * 6A | 5 | 26.3 | 4 | 11.7 | 6 | 30.0 | |
| NAT2 * 6B | 0 | 0 | 1 | 2.9 | 1 | 5.0 | |
| NAT2 * 7B | 0 | 0 | 1 | 2.9 | 0 | 0 |
NAT2 (Arylamine N-Acetyltransferase), PLWHA (People living with HIV/AIDS).
Thirteen alleles derived from these 7 common SNPs were verified, the majority of which (7/13) were classified phenotypically as slow acetylators. The others (5/13) were classified as rapid acetylators. There was no statistically significant difference between the groups studied. The most frequent allele observed with rapid acetylation was NAT2*4 (86%), followed by NAT2*11A (7%). With regard to slow acetylation, the most frequent allele was NAT2*5B (38%), followed by NAT2*5D (25%) and NAT2*6A (21%). The NAT2 allele frequencies of both the healthy control subjects and PLWHA/TB were very similar and were in Hardy-Weinberg equilibrium (Table 2).
In the NAT2*14A, an absence was observed of the AA genotype variant, both in the studied population and the control population. Within this locus, the most frequent genotype was GG (89.6%). For the NAT2*13A, the frequency of genotypes CC (43.3%) and CT (46.3%) was similar between the healthy and control groups. In the NAT2*6B, there was a genotypic frequency of 58.4% GG, GA 35.3% and 6.3% AA. Amongst the 173 individuals analyzed, only one individual presented the AA genotype variant in the NAT2*7A.
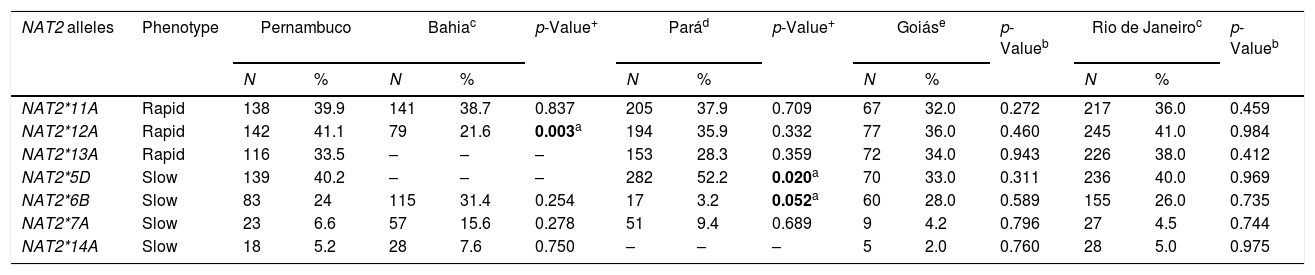
The frequencies of the variant NAT2 allele amongst individuals from different Brazilian regions are summarized in Table 3. In this cohort, the allele frequencies of the study population were different to those encountered in the Pará and Bahia populations. The main difference in the Pará population was in variant allele NAT2*6B. The frequency of this allele in the present study was 24%, while in the Pará population the frequency of the same allele was 3.2%. Another allele, NAT2*5D, was more frequent in the Pará population (52%) when compared to the frequency of same allele in our population (40%), with a statistically significant difference (p=0.020). The frequency of the variant allele NAT2*12A in the Bahia population was lower (21.6%) than that observed in this population (41.1%), also with a statistically significant difference (p=0.003).
Frequencies of the NAT2 alleles in individuals from different Brazilian regions.
| NAT2 alleles | Phenotype | Pernambuco | Bahiac | p-Value+ | Parád | p-Value+ | Goiáse | p-Valueb | Rio de Janeiroc | p-Valueb | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | ||||||
| NAT2*11A | Rapid | 138 | 39.9 | 141 | 38.7 | 0.837 | 205 | 37.9 | 0.709 | 67 | 32.0 | 0.272 | 217 | 36.0 | 0.459 |
| NAT2*12A | Rapid | 142 | 41.1 | 79 | 21.6 | 0.003a | 194 | 35.9 | 0.332 | 77 | 36.0 | 0.460 | 245 | 41.0 | 0.984 |
| NAT2*13A | Rapid | 116 | 33.5 | – | – | – | 153 | 28.3 | 0.359 | 72 | 34.0 | 0.943 | 226 | 38.0 | 0.412 |
| NAT2*5D | Slow | 139 | 40.2 | – | – | – | 282 | 52.2 | 0.020a | 70 | 33.0 | 0.311 | 236 | 40.0 | 0.969 |
| NAT2*6B | Slow | 83 | 24 | 115 | 31.4 | 0.254 | 17 | 3.2 | 0.052a | 60 | 28.0 | 0.589 | 155 | 26.0 | 0.735 |
| NAT2*7A | Slow | 23 | 6.6 | 57 | 15.6 | 0.278 | 51 | 9.4 | 0.689 | 9 | 4.2 | 0.796 | 27 | 4.5 | 0.744 |
| NAT2*14A | Slow | 18 | 5.2 | 28 | 7.6 | 0.750 | – | – | – | 5 | 2.0 | 0.760 | 28 | 5.0 | 0.975 |
NAT2 (Arylamine N-Acetyltransferase).
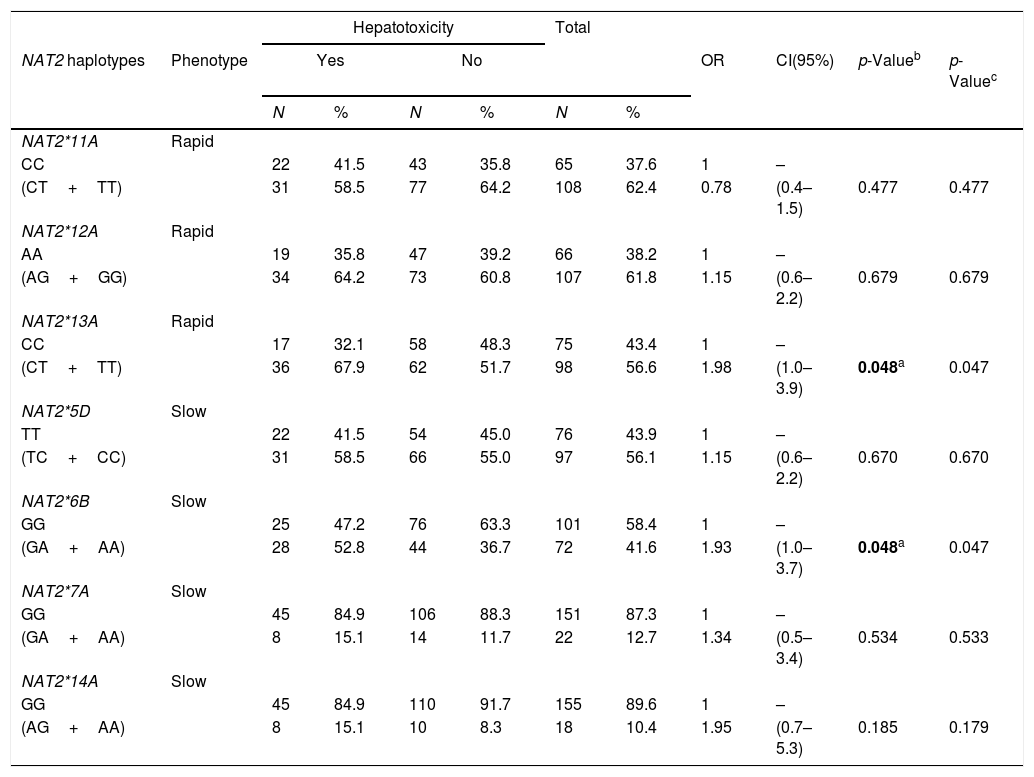
The hypothesis that PLWHA and TB patients encoding NAT2 variant alleles would be more likely to develop hepatotoxicity during TB treatment was tested. In order to perform such an analysis, the PLWHA and TB group was stratified taking into account the clinical outcomes of hepatotoxicity and the potential correlation was observed between the seven SNPs and the clinical outcome. It was found that the presence of the variant allele of both NAT2*13A (OR=1.98, 95%CI (1.0–3.9), p=0.048) and NAT2*6B (OR=1.93, 95%CI (1.0–3.7), p=0.048) constituted isolated risk factors for developing hepatotoxicity during TB treatment in PLWHA (Table 4).
Association of NAT2 genotypes with hepatotoxicity in PLWHA with tuberculosis. Recife, Pernambuco, Brazil.
| Hepatotoxicity | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NAT2 haplotypes | Phenotype | Yes | No | OR | CI(95%) | p-Valueb | p-Valuec | ||||
| N | % | N | % | N | % | ||||||
| NAT2*11A | Rapid | ||||||||||
| CC | 22 | 41.5 | 43 | 35.8 | 65 | 37.6 | 1 | – | |||
| (CT+TT) | 31 | 58.5 | 77 | 64.2 | 108 | 62.4 | 0.78 | (0.4–1.5) | 0.477 | 0.477 | |
| NAT2*12A | Rapid | ||||||||||
| AA | 19 | 35.8 | 47 | 39.2 | 66 | 38.2 | 1 | – | |||
| (AG+GG) | 34 | 64.2 | 73 | 60.8 | 107 | 61.8 | 1.15 | (0.6–2.2) | 0.679 | 0.679 | |
| NAT2*13A | Rapid | ||||||||||
| CC | 17 | 32.1 | 58 | 48.3 | 75 | 43.4 | 1 | – | |||
| (CT+TT) | 36 | 67.9 | 62 | 51.7 | 98 | 56.6 | 1.98 | (1.0–3.9) | 0.048a | 0.047 | |
| NAT2*5D | Slow | ||||||||||
| TT | 22 | 41.5 | 54 | 45.0 | 76 | 43.9 | 1 | – | |||
| (TC+CC) | 31 | 58.5 | 66 | 55.0 | 97 | 56.1 | 1.15 | (0.6–2.2) | 0.670 | 0.670 | |
| NAT2*6B | Slow | ||||||||||
| GG | 25 | 47.2 | 76 | 63.3 | 101 | 58.4 | 1 | – | |||
| (GA+AA) | 28 | 52.8 | 44 | 36.7 | 72 | 41.6 | 1.93 | (1.0–3.7) | 0.048a | 0.047 | |
| NAT2*7A | Slow | ||||||||||
| GG | 45 | 84.9 | 106 | 88.3 | 151 | 87.3 | 1 | – | |||
| (GA+AA) | 8 | 15.1 | 14 | 11.7 | 22 | 12.7 | 1.34 | (0.5–3.4) | 0.534 | 0.533 | |
| NAT2*14A | Slow | ||||||||||
| GG | 45 | 84.9 | 110 | 91.7 | 155 | 89.6 | 1 | – | |||
| (AG+AA) | 8 | 15.1 | 10 | 8.3 | 18 | 10.4 | 1.95 | (0.7–5.3) | 0.185 | 0.179 | |
NAT2 (Arylamine N-Acetyltransferase). OR (Odds ratio). CI (95%): confidence interval. PLWHA (People living with HIV/AIDS).
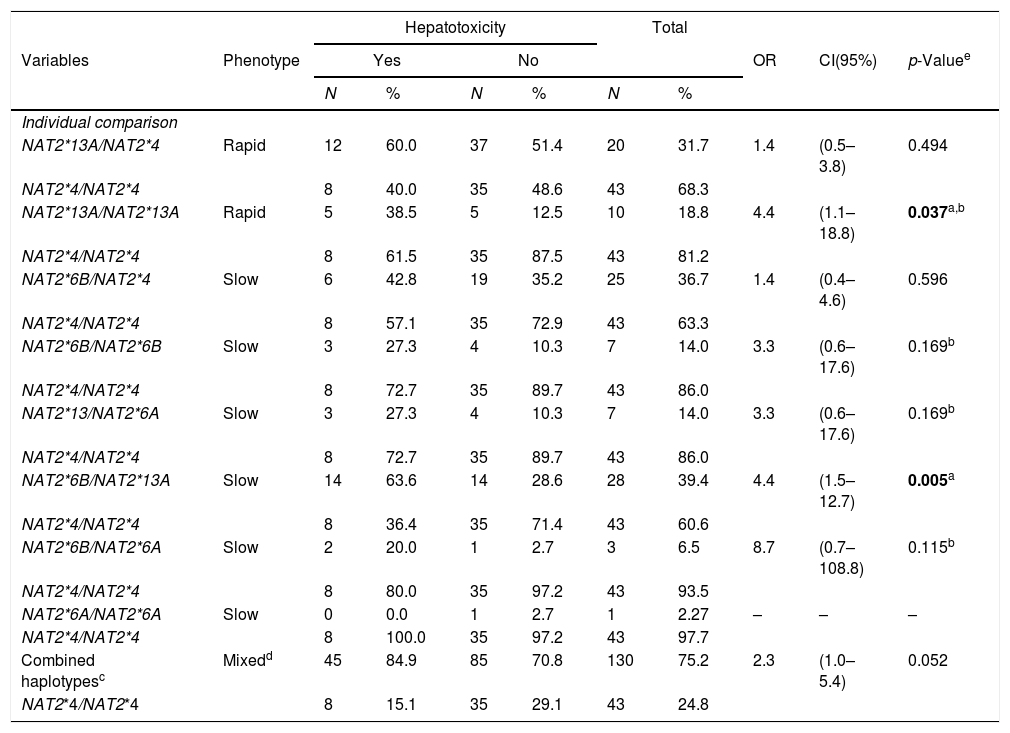
In the population studied, it was possible to define four haplotypes, [NAT2*4 (GC), NAT2*13A (GT), NAT2*6B (AC), NAT2*6A (AT)], with 8 genotype variants, [NAT2*4/NAT2*13A (GC/GT), NAT2*4/NAT2*6B (GC/AC), NAT2*13A/NAT2*13A (GT/GT), NAT2*13A/NAT2*6A (GT/AT), NAT2*13A/NAT2*6B (GT/AC), NAT2*6B/NAT2*6B (AC/AC), NAT2*6B/NAT2*6A (AC/AT) and NAT2*6A/NAT2*6A (AT/AT)], and one wild type [NAT2*4/NAT2*4 (GC/GC)] (Table 4).
In turn, the individual comparison between the wild type and each genotype containing allele variants indicated that the risk of developing hepatotoxicity during TB treatment in individuals with HIV/AIDS and signatures NAT2*13A/NAT2*13A (GT/GT) and NAT2*13A/NAT2*6B (GT/AC) was 4.4 times greater, and was statistically significant [p=0.037; (95% CI 1.1–18.8) and p=0.005; (95% CI 1.5–12.7), respectively]. A borderline association of a risk for developing hepatotoxicity was demonstrated in the analysis between combined haplotypes involving the variant group and the wild type (OR=2.3 (95% CI 1.0–5.4), p=0.052) (Table 5).
Association of NAT2 haplotypes with hepatotoxicity in PLWHA with tuberculosis. Recife, Pernambuco, Brazil.
| Hepatotoxicity | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Phenotype | Yes | No | OR | CI(95%) | p-Valuee | ||||
| N | % | N | % | N | % | |||||
| Individual comparison | ||||||||||
| NAT2*13A/NAT2*4 | Rapid | 12 | 60.0 | 37 | 51.4 | 20 | 31.7 | 1.4 | (0.5–3.8) | 0.494 |
| NAT2*4/NAT2*4 | 8 | 40.0 | 35 | 48.6 | 43 | 68.3 | ||||
| NAT2*13A/NAT2*13A | Rapid | 5 | 38.5 | 5 | 12.5 | 10 | 18.8 | 4.4 | (1.1–18.8) | 0.037a,b |
| NAT2*4/NAT2*4 | 8 | 61.5 | 35 | 87.5 | 43 | 81.2 | ||||
| NAT2*6B/NAT2*4 | Slow | 6 | 42.8 | 19 | 35.2 | 25 | 36.7 | 1.4 | (0.4–4.6) | 0.596 |
| NAT2*4/NAT2*4 | 8 | 57.1 | 35 | 72.9 | 43 | 63.3 | ||||
| NAT2*6B/NAT2*6B | Slow | 3 | 27.3 | 4 | 10.3 | 7 | 14.0 | 3.3 | (0.6–17.6) | 0.169b |
| NAT2*4/NAT2*4 | 8 | 72.7 | 35 | 89.7 | 43 | 86.0 | ||||
| NAT2*13/NAT2*6A | Slow | 3 | 27.3 | 4 | 10.3 | 7 | 14.0 | 3.3 | (0.6–17.6) | 0.169b |
| NAT2*4/NAT2*4 | 8 | 72.7 | 35 | 89.7 | 43 | 86.0 | ||||
| NAT2*6B/NAT2*13A | Slow | 14 | 63.6 | 14 | 28.6 | 28 | 39.4 | 4.4 | (1.5–12.7) | 0.005a |
| NAT2*4/NAT2*4 | 8 | 36.4 | 35 | 71.4 | 43 | 60.6 | ||||
| NAT2*6B/NAT2*6A | Slow | 2 | 20.0 | 1 | 2.7 | 3 | 6.5 | 8.7 | (0.7–108.8) | 0.115b |
| NAT2*4/NAT2*4 | 8 | 80.0 | 35 | 97.2 | 43 | 93.5 | ||||
| NAT2*6A/NAT2*6A | Slow | 0 | 0.0 | 1 | 2.7 | 1 | 2.27 | – | – | – |
| NAT2*4/NAT2*4 | 8 | 100.0 | 35 | 97.2 | 43 | 97.7 | ||||
| Combined haplotypesc | Mixedd | 45 | 84.9 | 85 | 70.8 | 130 | 75.2 | 2.3 | (1.0–5.4) | 0.052 |
| NAT2*4/NAT2*4 | 8 | 15.1 | 35 | 29.1 | 43 | 24.8 | ||||
OR (Odds ratio), CI (95%): confidence interval, PLWHA (People living with HIV/AIDS).
The metabolic pathway of isoniazid has been the basis for most of the recent studies seeking to explain the pathogenic mechanism of liver damage induced by the use of antituberculosis drugs, and the NAT2 slow acetylator phenotype has been documented as a strong risk factor for the occurrence of such liver damage [16,17]. In effect, although some studies have indicated different opinions, i.e., that rapid acetylators would be more vulnerable to liver damage induced by antituberculosis drugs due to the increased production of hepatotoxins, resulting from the rapid NAT2 enzyme activity, and that there is no relationship between the acetylation profile and the hepatotoxicity induced by these drugs [18,19], it is nonetheless believed that individuals phenotypically classified as slow acetylators accumulate more acetyl hydrazine, since it competes with the toxic metabolite of isoniazid by acetylation [20].
The NAT2 is located on chromosome 8, region 8p22, where seven SNPs are commonly identified in its coding region, in different world populations (<http://nat.mbg.duth.gr>).
The distribution and frequency of the alleles that encode the NAT2 may vary considerably between different ethnic groups. The NAT2*14A is more common in native African populations and African Americans. However, it is virtually absent in the East. The frequencies of the NAT2*13A and NAT2*6B are similar in most populations studied throughout the world. In the case of NAT2*5D and NAT2*7A, there was a greater frequency amongst Moroccans, Amerindians and Asians. NAT2*11A and NAT2*12A are rare in most populations [8,13,21–24].
The Brazilian population is made up of an extensive mix of three different ancestral roots: Amerindians, Europeans and Africans, which makes it one of the most heterogeneous populations in the world [25,26]. The first Brazilian studies describing the genotypic profile of NAT2 were conducted in the states of Rio de Janeiro and Goiás where degrees of variation in the genotypic profile of NAT2 were demonstrated [11–27]. The allele frequencies observed in our cohort were similar to those found in the populations of Rio de Janeiro and Goiás, although they were different from the populations of the states of Pará and Bahia [27–29]. These data reinforce the need for further studies in order to address this issue throughout the different regions of the country so that the genotypic profile of the NAT2 in Brazil may be better explained. In other Brazilian studies the data were restricted to reporting the acetylation phenotypic profile and therefore, the allele and genotype frequencies were not presented [4,5,30,31]. In this present approach, for all the studied SNPs, were observed that the allele and genotype frequencies of the healthy control population were similar to those observed in the test population (PLWHA). Thus, we may affirm the good representation of the cohort for the state of Pernambuco.
The presence of the variant allele in the NAT2*13A (282T) and NAT2*6B (590A) loci was an independent factor associated with elevated aminotransferase levels after initiating antituberculosis drugs in PLWHA [(OR=1.98, 95%CI 1.0–3.9, p=0.048) and (OR=1.93, 95%CI 1.0–3.7, p=0.048), respectively]. The NAT2*6B is a non-synonymous polymorphism that involves the replacement of amino acid residue 197 Arg-Gln (arginine-glutamine) with a consequent decrease in the acetylation capacity, due to a change in the catalytic site, and which therefore justifies our findings [32]. Poor individual metabolizers present low enzymatic efficiency of NAT acetyltransferase_2, and thereby allow more time for the drug to circulate in the body. As a result, the toxicity produced by the drug also becomes higher [33]. In contrast to this, the NAT2*13A is a synonymous polymorphism, i.e., its presence alone does not prompt a change in the peptide composition, and is also unable to change the slow/rapid molecular phenotype acetylator in patients [34], diverging from the risk association for hepatotoxicity associated with the NAT2*13A in PLWHA, as observed in the present study. One recent publication on genetic context, could be a key element in explaining the phenotype definition in genetically influenced diseases and the contradictory effects of individual genetic markers (SNPs) that have been observed in different populations [35].
The special conditions concerning two different coinfecting pathogens, with concomitant treatment against HIV and TB, and the potential interaction between them, represent important confounding factors for the outcome of hepatotoxicity. Indeed, it has recently been described that the properties of using fluconazole, being malnourished and the presence of a NAT2 slow acetylator profile were each isolated risk factors for developing hepatotoxicity. It is suspected that hepatitis B and C viruses and alcohol consumption may be associated with increased hepatotoxicity in PLWHA during tuberculosis therapy, although in this previous study this association was not observed [4].
In the present study, it has been verified that the BMI (p=0.002) and CD4 (p=0.022) were statistically associated with hepatotoxicity, suggesting that these two variables were likely to be confounding factors. Thus, the analysis was undertaken of these two variables with the SNPs and it was verified that they presented no statistically significant difference. This fact interferes with the definition of confounding factor. It is therefore our belief that these two variables should not be used to adjust the model in logistic regression analysis. It is a fact that drug interactions modify the response to pharmacotherapy and are also a risk for drug toxicity. While on the one hand, the use of HAART is associated with a higher risk of hepatotoxicity, because it potentiates the hepatotoxic effect of the antituberculosis drugs, on the other, we also believe that the interaction between anti-HIV and antituberculosis drugs probably has no influence over the association between NAT2 variants and hepatotoxicity. Although most of the study population (78.6%) used HAART, of whom 33.9% presented with hepatotoxicity during the first 60 days of TB treatment, no statistically significant difference was observed between the use of HAART and the occurrence of hepatotoxicity. We chose to analyze the combination of ARV classes due to the wide variation in the antiretroviral drugs used by the population studied. Certainly, if we had analyzed the association of each drug with the outcome, the power of the study would have been limited.
Finally, the present study has the limitations of an observational study with PLWHA attending a routine medical care setting. The exposures related to drugs registered by the attending physician were studied. We did not investigate the concomitant use of medicinal herbs in this population.
The findings of this study have demonstrated that providing greater support for PLWHA undergoing treatment for tuberculosis may well contribute to greater control in managing the drugs used by this population, especially those patients with genotypes NAT2*13A/NAT2*13A and NAT2*13A/NAT2*6B.
In conclusion, this study suggests that NAT2*13A and NAT2*6B variant alleles are risk factors for developing hepatotoxicity, and that PLWHA with genotypes NAT2*13A/NAT2*13A and NAT2*13A/NAT2*6B present a higher chance of developing hepatotoxicity during tuberculosis treatment. We believe that in the near future, the treatment of tuberculosis should become individualized, namely, drugs and dosages should be adapted according to the NAT2 phenotypic profile of each patient. Further studies with different populations and larger sample sizes are required to verify the universality of our findings.AbbreviationsAIDS acquired immunodeficiency syndrome alanine aminotransferase arginine glutamine antiretroviral therapy aspartate aminotransferase body mass index base pairs lymphocyte cell with cell membrane surface protein 4 confidence interval cytochrome P450 2E1 deoxyribonucleic acid deoxynucleotides glutathione S-transferase highly active antiretroviral therapy human immunodeficiency virus water magnesium chloride manganese superoxide dismutase arylamine N-acetyltransferase non-nucleoside reverse transcriptase inhibitor nucleoside reverse transcriptase inhibitor odds ratio polymerase chain reaction people living with HIV/AIDS protease inhibitor/ritonavir rifampicin, isoniazid, pyrazinamide rifampicin, isoniazid, pyrazinamide, ethambutol single nucleotide polymorphisms thermus aquaticus polymerase tuberculosis World Health Organization
All authors contributed equally to this article and all authors reviewed and approved the manuscript prior to submission.
Financial supportThis study received financial support from the Brazilian Health Ministry/DST/AIDS/UNESCO (CSV Project 182/06) and the Ministry of Science and Technology CNPq/MSSCTIE-DECIT (Case N°10567/2006-0). The following authors received partial support from CNPq: RAAX – Productivity Grant 308311/2009-4; MFPMA – Productivity Grant 301779/2009-0; and HRL – Study Grant 310911/2009-5.
Ethical aspectsThis study was approved by the Research Ethics Committee at the Universidade Federal de Pernambuco, Brazil, under protocol No. 254/05. All participants in the study signed an Informed Consent Form, as required by the statement on ethics contained in Resolution N°466/12 of the Brazilian National Health Council.
Conflict of interestThe authors have no conflicts of interest to declare.