Special issue on hepatocellular carcinoma (HCC) and hepatitis B and C as its main causes worldwide
Más datosThis study aimed to investigate miR-3682 as a biomarker in hepatocellular carcinoma (HCC).
Materials and methodsMiRNA and RNA profiles of 375 HCC tissues and 50 normal liver samples were downloaded from The Cancer Genome Atlas (TCGA) database. Multivariate Cox regression and Kaplan-Meier analyses were applied to examine the prognostic value of factors. Target genes of miR-3682 were analyzed by TargetScan and dual-luciferase reporter assay. Online Database for Annotation, Visualization, and Integrated Discovery (DAVID) to perform KEGG pathway enrichment. Cell counting kit-8, colony formation and migration and invasion assays were performed to analyze biological behaviors of HCC cells.
ResultsMiR-3682 was identified to be highly expressed in HCC tissues and cell lines. And miR-3682 was negatively and independently associated with the outcome of HCC patients. Inhibition of miR-3682 suppressed HCC cell viability and mobility. ADRA1A, predicted and confirmed as the novel target of miR-3682, was an independent and positive prognostic predictor for HCC. In addition, the knockdown of ADRA1A partially offset the inhibitory effect of miR-3682 inhibitor on the growth and mobility of HCC cells. DAVID enrichment and western blot of key signaling-related proteins analyses revealed that miR-3682 inactivated 5′-AMP-activated protein kinase (AMPK) signaling by negatively regulating ADRA1A. Mechanically, it was partially through suppressing AMPK signaling via targeting ADRA1A that miR-3682 supported the HCC cell malignant phenotype.
ConclusionsThis study implicates that miR-3682 plays an oncogenetic role in HCC and can be considered a novel therapeutic target and prognostic indicator of HCC.
With approximately 42,810 new cases and 30,160 deaths in the United States, liver hepatocellular carcinoma (HCC) is estimated to claim the fifth-largest cancer death in 2020 [1]. The main etiology of HCC is clarified as constant hepatitis virus infection [2]. However, due to late diagnosis and a high rate of recurrence and metastasis, HCC has a quite dismal outcome with a 5-year survival of < 18% [3]. Therefore, it is urgently needed to identify new biomarkers for effective treatments and more satisfying outcomes of HCC patients. This brings with the requirement to explore molecular mechanisms governing the carcinogenesis of HCC.
MicroRNAs (miRNAs), a family of short non-coding RNAs, can bind to 3′ untranslated region (3’-UTR) of mRNAs to degrade or post-translationally inhibit the expression of target genes [4]. MiRNAs are reported to participate in the processes of cell proliferation, apoptosis and mobility [5]. Many studies have revealed that miRNAs exert critical roles in the progression of HCC. Shen et al. reviewed that several miRNAs can promote HCC cell proliferation or mobility via regulating downstream genes and relevant signaling pathways, including p53, Glucocorticoids and HGF signaling pathways [6]. Abnormal expression of some oncogenetic miRNAs including miR-1275 [7] and miR-155 [8] and some anti-tumor miRNAs including miR-486-5p [9] and miR-34a [10] were detected during HCC progression. Besides, multiple miRNAs were identified as promising therapeutic and prognostic biomarkers for HCC [11]. All the results highlighted the crucial biological roles of miRNAs in HCC. Importantly, the expression of miR-3682 was revealed to be elavated in colon cancer samples and facilitated cancer cell proliferation and migration [12]. Moreover, in this study, we detected that miR-3682 was also highly expressed in HCC and associated with a poor outcome of HCC. Given the previous and our findings, we further explore the biological roles of miR-3682 in HCC.
In our study, miR-3682 was revealed to be up-regulated in HCC samples and significantly associated with the death of HCC patients. MiR-3682 was negatively and independently correlated with the prognosis of HCC patients. Further, we identified α1A- Adrenergic receptor (ADRA1A) as a novel target of miR-3682 and miR-3682 exerted a supportive role in HCC cell biological behaviors via targeting ADRA1A. Our study suggests miR-3682 as an oncogenic miRNA for HCC and a promising therapeutic application of miR-3682 in HCC.
2Materials and methods2.1Bioinformatics analysisMiRNA and mRNA sequencing data, including 375 HCC tissues and 50 normal liver samples, were obtained from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/). Genes were subjected to the online Database for Annotation, Visualization, and Integrated Discovery (DAVID; v.6.8; http://david.ncifcrf.gov/tools.jsp) to perform Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment. In addition, the online bioinformatics tool TargetScan was used to forecast the downstream targets of miR-3682.
2.2Cell cultureHCC cell lines, including HepG2 and Huh7 were purchased from American Type Culture Collection (ATCC, USA). And human liver immortal cell line L-02 was from China Center for Type Culture Collection (CCTCC, China). All the cell lines were cultured in RPMI 1640 (Invitrogen, USA) containing 10% FBS in a CO2 incubator.
2.3Cell transfectionWhen the confluence reached 80%, cells were transfected mediating with Lipofectamine2000 (Thermo Fisher Scientific, USA) according to the manufacture's direction. Cells were transfected with si-ADRA1A (5’- CCUUUCAGAAUGUCUUGAG -3’), si-negative control(NC) (5’- AAUUCUCCGAACGUGUCACGU -3’), miR-3682 inhibitor, mimic or NC which were synthesized by Genechem (Shanghai, China). After 24 h transfection, cells were collected for further analysis.
2.4RNA isolation and Quantitative real-time PCR (RT-qPCR)According to the manufacturer's directions, total RNAs were isolated from HepG2, Huh7 and L-02 cells using TRIzol reagent (Thermo Scientific, USA). Complementary DNAs (cDNAs) were reversely transcribed from the isolated RNAs and amplified using the following primers listed in Table 1 and KiCqStart One Step Probe RT-qPCR Readymix (Sigma-Aldrich, USA). RT-qPCR was performed on StepOnePlus Real-Time PCR System (Life Technologies, USA). The expression of miR-3682 and ADRA1A was measured by SYBR Green Quantitative RT-PCR Kit (Merck Millipore, German) with 2−ΔΔCT method and normalized by U6 and GAPDH, respectively.
2.5Western BlotCollected cells were incubated in RIPA buffer for 30 min. Cell lysates were centrifuged at 21,000 g for 15 min to collect the supernate. The proteins precipitated from the supernate were dissolved in 1X loading buffer, maintained at 95 ℃ for 5 min followed by separation with 10% SDS-PAGE. Then the separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes, which was subsequently blocked in 5% non-fat milk. After the incubation with the following primary antibodies overnight at 4 ℃, the membrane was washed, followed by the maintenance with appropriate secondary antibodies for 1 h at room temperature. BeyoECL Star (Beyotime, China) was used to react with secondary antibodies to visualize protein bands. And signal intensity was determined on an ImageQuant LAS 4000 system (GE Healthcare). GAPDH was used to normalize protein expression. The primary antibodies were as follows: anti- ADRA1A antibody at 1:1000, ab137123, Abcam, UK; anti- GAPDH antibody at 1:1000, ab8245, Abcam, UK; anti- Phospho-AMPKα (Thr172) antibody at 1:1000, #50081, Cell Signaling Technology, USA; anti- AMPKα antibody at 1:1000, #2532, Cell Signaling Technology, USA; anti- Phospho-mTOR (Ser2448) antibody at 1:1000, ab109268, Abcam, UK; anti- mTOR antibody at 1:1000, ab134903, Abcam, UK.
2.6Cell counting kit-8 assayAfter transfection, 100 μL of cell suspension (10 cells/μL) was planted into a 96-well plate. After 0, 24,48 or 72 h transfection, 10 μL cell counting kit-8 reagent (CCK-8, Beyotime Biotechonology, China) was added into the medium and the cells were cultured for another 1.5 h at 37 ℃. And the OD450 values were measured using a microplate reader.
2.7Cell clonogenic assayThe transfected cells were implanted in a 6-well plate (400 cells/well). The cells were cultured until the macroscopic colonies appeared. Then the cells were fixed with 6% (v/v) glutaraldehyde and stained with 0.1% (w/v) crystal violet. The number of the formed colonies were counted using an optical microscope.
2.8Transwell migration and invasion assayFor invasion assay, the top chamber was firstly coated with matrigel. The 100 μL cell suspension in FBS-free medium was added to the coated top insert, and 500 μL complete medium was added to the lower insert as attractant. Then the chamber was incubated at 37 ℃ for 24 h. The invasive cells were fixed with 6% (v/v) glutaraldehyde and stained with 0.1% (w/v) crystal violet. Then cells in five random fields of view were counted under a microscope. For migration assay, the top insert was not pre-coated with matrigel, and the other steps were the same as those in invasion assay.
2.9Dual-luciferase reporter assayThe complementary sites between miR-3682 and 3’-UTR of ADRA1A were predicted by TargetScan. The wild-type (WT) or mutant (MUT) 3’-UTR of ADRA1A was cloned into the pGL3-control luciferase reporter vector (Promega, USA) and then co-transfected into HepG2 and Huh7 cells with miR-3682 inhibitor/mimic/mimic NC mediated by Lipofectamine2000 (Thermo Scientific, USA). The Dual-Luciferase Reporter Assay system (Promega) was utilized to quantify the luciferase activity.
2.10Statistics analysisSPSS 22.0 (IBM, USA) and GraphPad Prism 7 (GraphPad Software, USA) were utilized to statistically analyze the data. Kaplan-Meier (KM) method with log-rank test was performed to analyze the correlation of miR-3682 and ADRA1A expression and the outcome of HCC patients. Student's t-test was used to compare between two independent groups and one-way analysis of variance was for three or more groups. Univariate and multivariate Cox regression was performed to analyze the association between the outcome of HCC patients and the clinical features and the expression of miR-3682 and ADRA1A, while Chi-square test was for the association between miR-3682 and ADRA1A expression and the clinicopathological features of HCC patients.
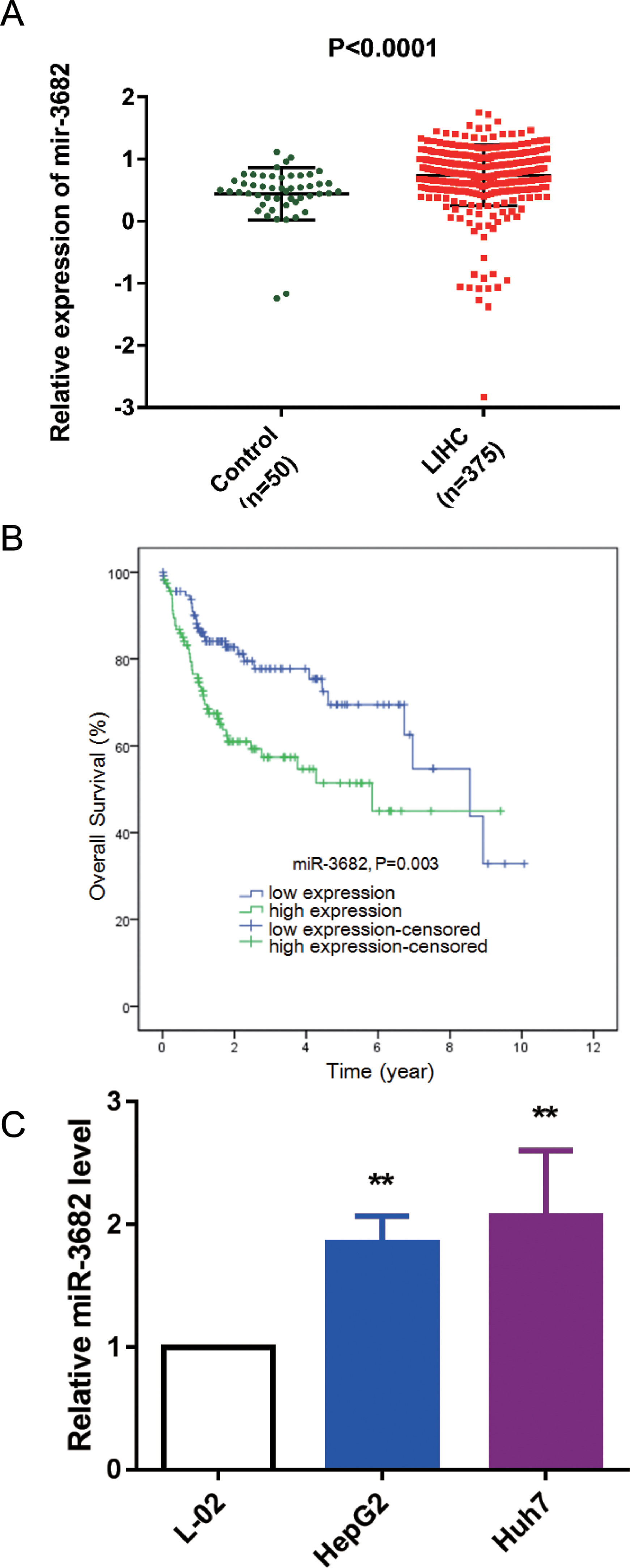
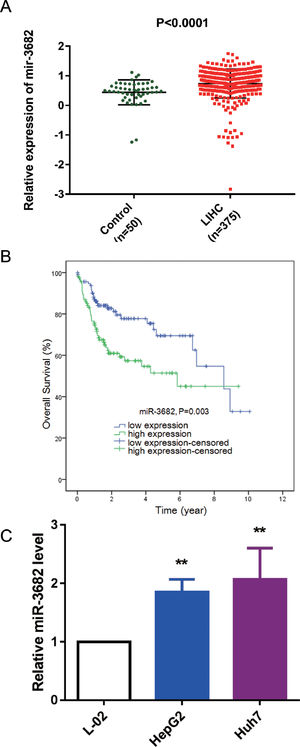
3Results3.1MiR-3682 was up-regulated in HCC samples and independently associated with a dismal outcome of HCC patientsTo uncover the roles of miR-3682 in HCC, we firstly analyzed miR-3682 expression in HCC data from TCGA database. As depicted in Fig. 1a, the level of miR-3682 expression was notably elevated in HCC tissues (n = 375) compared to the non-cancerous samples (n = 50, P < 0.0001). Chi-square test was used to evaluate the relationship between miR-3682 expression and clinical parameters of HCC patients. We found there was a notable association between miR-3682 expression and the death of HCC patients (Table 2, P < 0.05). Next, HCC patients with higher expression of miR-3682 exhibited a worse outcome than those of miR-3682 low expression group (Fig. 1b, P = 0.003). Further, univariate and multivariate Cox regression uncovered that miR-3682 could be considered as an independent factor of HCC outcome (Table 3, P < 0.01). In addition, we analyzed miR-3682 expression in HCC cells and normal human liver cells. The results showed that miR-3682 was highly expressed in HCC cells including HepG2 and Huh7 cells compared with normal L-02 cells (Fig. 1c, P < 0.01). Collectively, miR-3682 expression was raised in HCC samples and could be considered as a potential and independent prognostic predictor of HCC patients.
MiR-3682 was up-regulated in HCC tissues and cells. (A) Expression of miR-3682 was notably higher in HCC tissues (n = 375) than that in the non-cancerous samples (n = 50). LIHC, liver hepatocellular carcinoma. The data are exhibited as mean±SD, P < 0.0001. (B) Kaplan-Meier curves revealed that the survival outcome of miR-3682 high expression group (n = 116) is worse than that of low expression group (n = 115). The 231 HCC patients with complete clinic information were separated into high and low expression groups with the cutoff value of miR-3682 median expression. (C) The miR-3682 expression was measured by RT-qPCR in HepG2, Huh7 and L-02 cells. The data are exhibited as mean±SD with triplicates. ⁎⁎P < 0.01
The association of miR-3682 expression with clinical features of HCC patients (n = 231) was analyzed by Chi-square test.
| Characteristics | Expression of miR-3682 | P value | |
|---|---|---|---|
| Low | High | ||
| Age | 0.100 | ||
| <60 | 54 | 67 | |
| ≥60 | 61 | 49 | |
| Gender | 0.127 | ||
| Female | 30 | 41 | |
| male | 85 | 75 | |
| Grade | 0.054 | ||
| G1+G2 | 71 | 57 | |
| G3+G4 | 44 | 59 | |
| Stage | 0.806 | ||
| I+II | 82 | 81 | |
| III+IV | 33 | 35 | |
| T-Stage | 0.691 | ||
| T1+T2 | 84 | 82 | |
| T3+T4 | 31 | 34 | |
| Node metastasis | 0.993 | ||
| N0 | 113 | 114 | |
| N1 | 2 | 2 | |
| Distant metastasis | 0.566 | ||
| M0 | 114 | 114 | |
| M1 | 1 | 2 | |
| Death | 0.018* | ||
| No | 87 | 72 | |
| Yes | 28 | 45 | |
The association between miR-3682 expression and clinical features and the prognosis of HCC patients were analyzed by univariate and multivariate Cox regression (n = 231).
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Pvalue | HR | 95%CI | Pvalue | HR | 95%CI | |
| miR-3682 expression (low/high) | 0.003* | 2.045 | 1.268-3.300 | 0.001* | 2.213 | 1.361-3.598 |
| Stage (I+II/ III+IV) | 0.000* | 3.129 | 1.972-4.966 | 0.529 | 1.896 | 0.259-13.897 |
| T-Stage (T1+T2/ T3+T4) | 0.000* | 3.147 | 1.982-4.996 | 0.596 | 1.715 | 0.233-12.619 |
| Distant metastasis (M0/M1) | 0.019* | 4.019 | 1.259-12.827 | 0.310 | 1.849 | 0.565-6.058 |
| Node metastasis (N0/N1) | 0.300 | 2.108 | 0.515-8.628 | |||
| Age (<60/≥60) | 0.405 | 1.216 | 0.767-1.927 | |||
| Gender (Female/Male) | 0.273 | 0.766 | 0.476-1.233 | |||
| Grade (G1+G2/G3+G4) | 0.765 | 1.073 | 0.676-1.703 | |||
HR: hazard ratio, CI: confidence interval
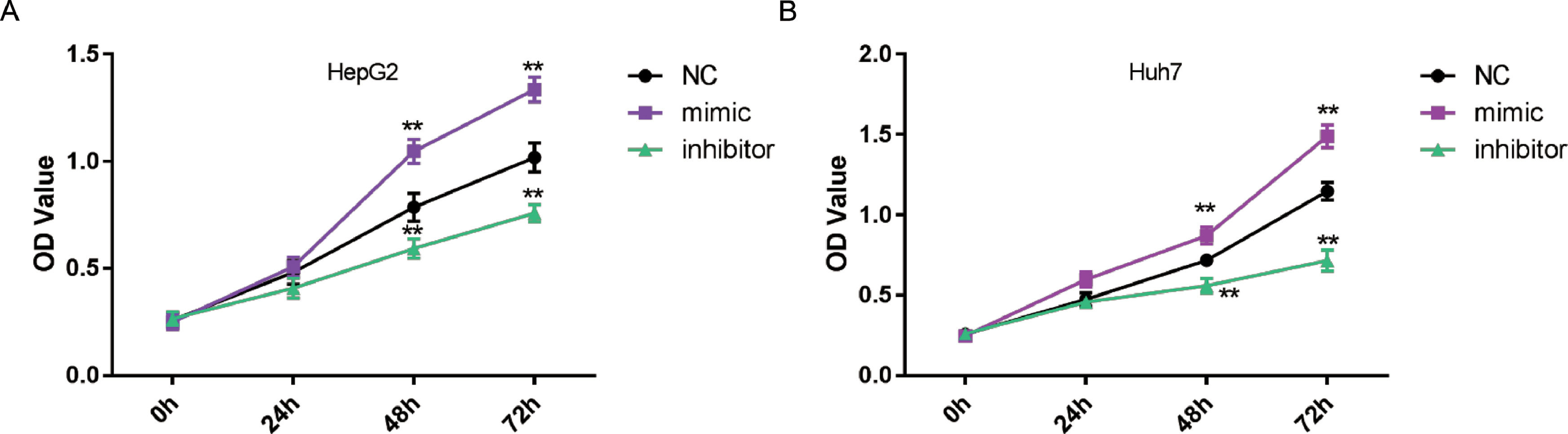
Next, CCK-8 assays were conducted to assess the role of miR-3682 in malignant growth of HCC cells. OD values at 450 nm were measured after 0,24,48 and 72 h transfection to quantify cell viability. As shown in Fig. 2a and b, miR-3682 mimic transfection remarkably augmented HepG2 and Huh7 cell proliferation, while miR-3682 inhibitor transfection hindered HCC cell viability compared to NC groups (P < 0.01). The results indicated that miR-3682 exerted a supportive effect on HCC cell growth.
MiR-3682 significantly promoted HCC cell viability. (A) The proliferation of HepG2 cells transfected with miR-3682 inhibitor, mimic or mimic NC was measured by CCK-8 assay. (B) The proliferation of Huh7 cells transfected with miR-3682 inhibitor, mimic or mimic NC was measured by CCK-8 assay. The data are exhibited as mean±SD with triplicates. ⁎⁎P < 0.01
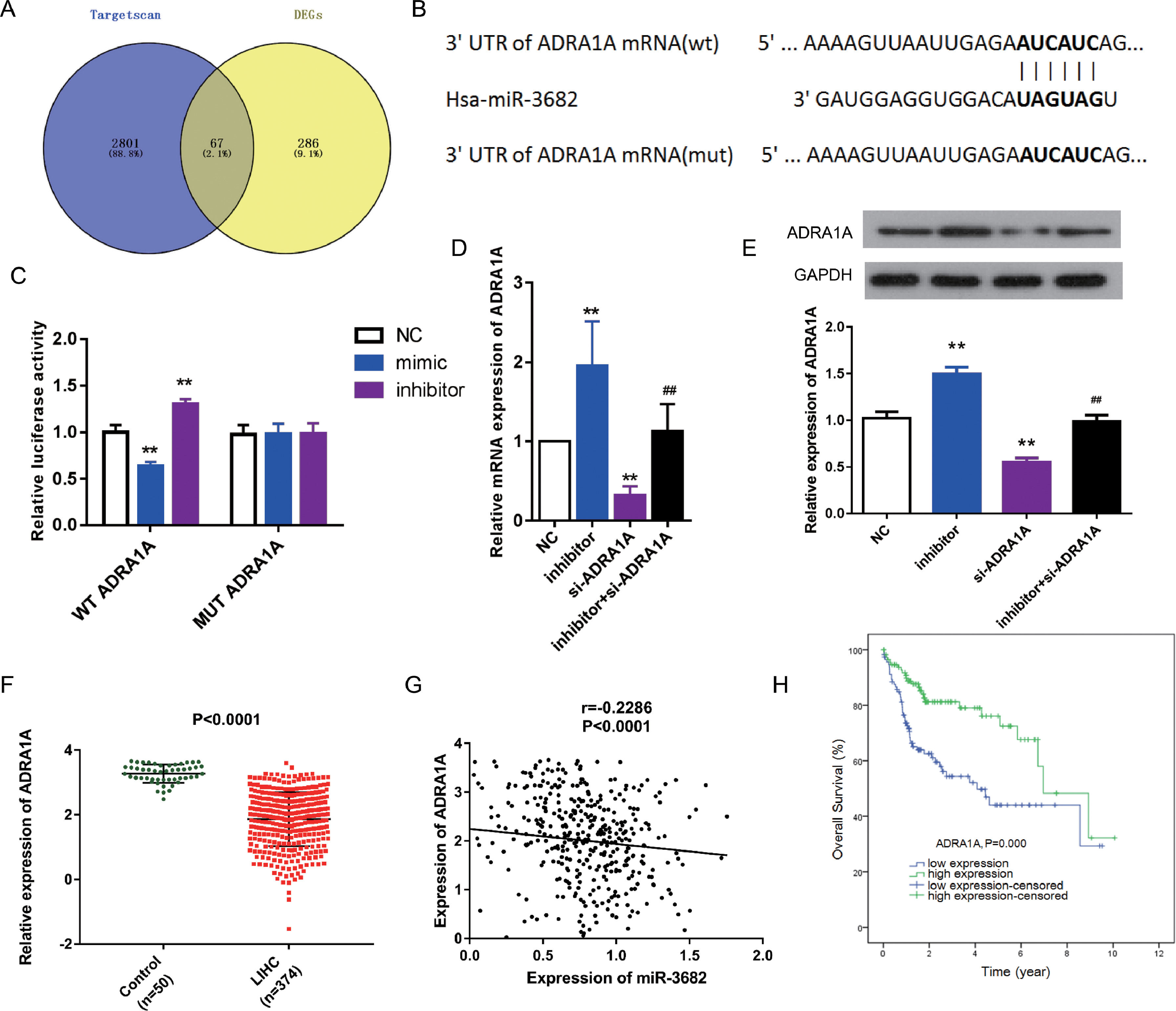
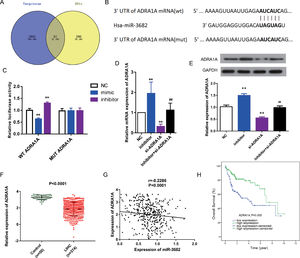
To uncover the mechanisms of how miR-3682 regulated malignancy of HCC cells, we further identified target genes of miR-3682. First, we analyzed the down-regulated genes in HCC samples. Subsequently, we compared the predicted downstream targets of miR-3682 using TargetScan with those down-regulated genes, and obtained 67 shared candidate target genes (Fig. 3a). The 67 genes were subjected to DAVID database to analyze the enriched KEGG pathways and four genes, namely ADRA1A, GYS2, FOXO1 and PPARGC1A, were enriched in one cancer-related pathway, 5′-AMP-activated protein kinase (AMPK) signaling pathway (Supplementary Table1). Due to the most evident down-regulation in HCC, ADRA1A was selected as the potential target of miR-3682. Next, we performed the following analysis to confirm the notion. We first performed dual-luciferase reporter assay to investigate the interaction of miR-3682 with ADRA1A. The complementary sites between miR-3682 and ADRA1A were forecasted by TargetScan (Fig. 3b). And 3’UTR of ADRA1A containing WT or MUT sequence was co-transfected into HCC cells with miR-3682 mimic, inhibitor or mimic NC. The luciferase activity following the co-transfection of miR-3682 mimic and WT ADRA1A was notably decreased compared with the NC group, while when miR-3682 inhibitor was co-transfected with WT ADRA1A, the luciferase activity was increased compared with the NC group (Fig. 3c, P < 0.01). Nonetheless, when MUT ADRA1A was co-transfected into HCC cells with miR-3682 mimic, inhibitor or mimic NC, there was no significant change in luciferase activity (Fig. 3c). The results reveal that miR-3682 can bind to the WT 3’-UTR of ADRA1A in luciferase reporter vector to hinder luciferase expression, whereas there was no interaction between miR-3682 and MUT 3’-UTR of ADRA1A. Besides, si- ADRA1A was co-transfected with miR-3682 inhibitor into HepG2 cells to investigate the effect of miR-3682 on ADRA1A expression. MiR-3682 inhibitor markedly augmented the mRNA and protein expression of ADRA1A compared with NC group, and the co-transfection of si- ADRA1A and inhibitor significantly elevated ADRA1A expression compared with si-ADRA1A group (Fig. 3d and e, P < 0.01). These results implicate that ADRA1A is a direct target of miR-3682.
ADRA1A was a direct target gene of miR-3682. (A) Venn diagram showed the number of the predicted target genes of miR-3682 (purple) and the down-regulated genes in HCC samples (yellow). (B) The binding sites in 3’-UTR of ADRA1A to miR-3682 were predicted by TargetScan and then artificially mutated. (C) HepG2 cells were co-transfected with miR-3682 inhibitor/NC and a luciferase reporter plasmids containing ADRA1A 3′-UTR WT/Mut, and the luciferase activity was measured. The data are exhibited as mean±SD with triplicates. ⁎⁎P < 0.01 vs. inhibitor NC group. ##P < 0.01 vs. inhibitor group. (D, E) The mRNA (D) and protein (E) expression of ADRA1A in HepG2 transfected with miR-3682 inhibitor/NC and/or si-ADRA1A was determined by RT-qPCR and western blot assays, respectively. The data are exhibited as mean±SD with triplicates. ⁎⁎P < 0.01 vs. inhibitor NC group. ##P < 0.01 vs. inhibitor group. (F) The expression of ADRA1A was measured in HCC tissues (n = 375) and the normal samples (n = 50). P < 0.0001. (G) The correlation analysis between ADRA1A and miR-3682 were analyzed by GraphPad Prism 7. R = -0.2286, P < 0.0001. (H) Kaplan-Meier curves revealed that the survival outcome of ADRA1A high expression group (n=114) was better than that of low expression group (n = 114). The 228 HCC patients with complete clinic information were separated into high and low expression groups with the cutoff value of ADRA1A median expression. P = 0.000
To further verify this notion, we measured the ADRA1A expression in HCC samples from TCGA database. ADRA1A was detected to be down-regulated in HCC tissues (n=374) compared to the normal samples (n = 50, Fig. 3f, P < 0.0001). In addition, the level of ADRA1A mRNA was negatively associated with the expression of miR-3682 (Fig. 3g, R = -0.2286, P < 0.0001). Furthermore, subsequent bioinformatics analysis indicated that ADRA1A exerted a clinical effect on HCC opposite to miR-3682. High ADRA1A expression was significantly associated with lower cancer stage and non-death status of HCC patients (Table 4, P < 0.05). KM curves indicated that HCC patients with ADRA1A high expression (n = 114) exhibited a better prognosis than ADRA1A low expression group (n = 114, Fig. 3h, P = 0.000). And multivariate Cox regression demonstrated that ADRA1A expression had a potential to be an independent risk factor of HCC prognosis (Table 5, P < 0.05). Collectively, these results legibly indicates that ADRA1A is a target of miR-3682 and plays an anti-tumorigenic role in HCC.
The association of ADRA1A expression with clinical features of HCC patients (n = 228) was analyzed by Chi-square test.
| Characteristics | Expression of ADRA1A | P value | |
|---|---|---|---|
| Low | High | ||
| Age | 0.691 | ||
| <60 | 60 | 57 | |
| ≥60 | 54 | 57 | |
| Gender | 0.444 | ||
| Female | 37 | 32 | |
| male | 76 | 82 | |
| Grade | 0.051 | ||
| G1+G2 | 58 | 72 | |
| G3+G4 | 56 | 41 | |
| Stage | 0.045* | ||
| I+II | 74 | 87 | |
| III+IV | 40 | 26 | |
| T-Stage | 0.103 | ||
| T1+T2 | 77 | 88 | |
| T3+T4 | 37 | 26 | |
| Node metastasis | 0.313 | ||
| N0 | 111 | 113 | |
| N1 | 3 | 1 | |
| Distant metastasis | 0.561 | ||
| M0 | 112 | 113 | |
| M1 | 2 | 1 | |
| Death | 0.001* | ||
| No | 65 | 89 | |
| Yes | 49 | 25 | |
The association between ADRA1A expression and clinical features and the prognosis of HCC patients were analyzed by univariate and multivariate Cox regression (n = 228).
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Pvalue | HR | 95%CI | Pvalue | HR | 95%CI | |
| ADRA1A expression (low/high) | 0.001* | 0.431 | 0.266-0.699 | 0.001* | 0.448 | 0.275-0.728 |
| Stage (I+II/ III+IV) | 0.000* | 2.921 | 1.846-4.622 | 0.812 | 1.275 | 0.173-9.402 |
| T-Stage (T1+T2/ T3+T4) | 0.000* | 2.954 | 1.866-4.677 | 0.440 | 2.200 | 0.297-16.314 |
| Distant metastasis (M0/M1) | 0.019* | 4.034 | 1.263-12.882 | 0.323 | 1.821 | 0.554-5.988 |
| Node metastasis (N0/N1) | 0.296 | 2.122 | 0.518-8.692 | |||
| Age (<60/≥60) | 0.329 | 1.256 | 0.794-1.987 | |||
| Gender (Female/Male) | 0.369 | 0.805 | 0.501-1.293 | |||
| Grade (G1+G2/G3+G4) | 0.898 | 1.031 | 0.648-1.640 | |||
HR: hazard ratio, CI: confidence interval
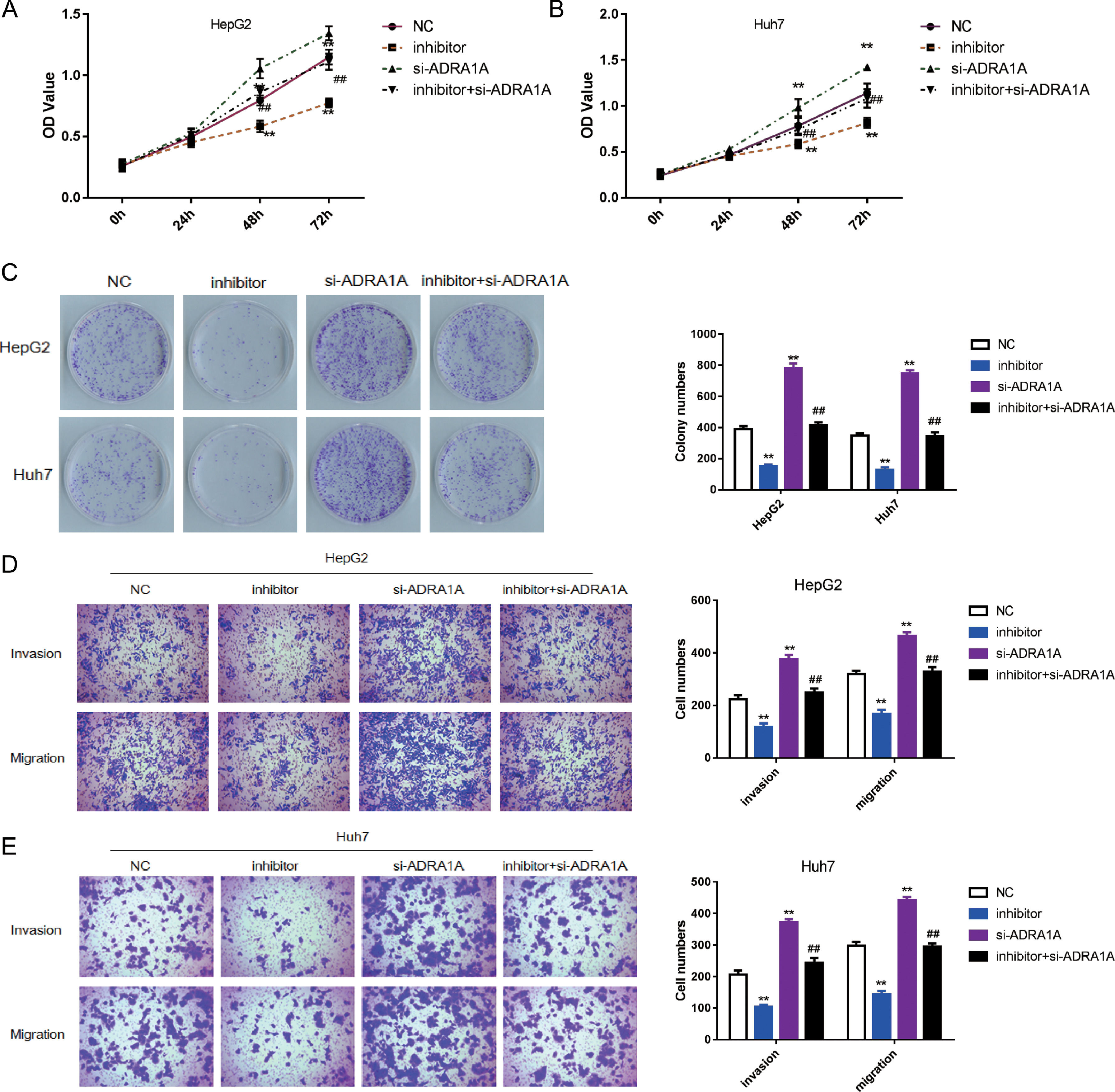
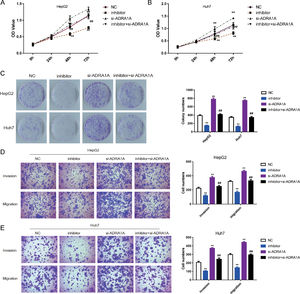
To explore whether ADRA1A participated in the regulation of malignant phenotype by miR-3682, we transfected HepG2 and Huh7 cells with si- ADRA1A and/or miR-3682 inhibitor to perform a rescue experiment and evaluated the phenotype of these cells. As depicted in Fig. 4 a–c, miR-3682 inhibitor lowered OD values and number of cell colonies significantly in HCC cells compared with the NC group; when HCC cells were transfected with ADRA1A siRNA, OD values and the number of cell colonies were increased notably compared with the NC group; when ADRA1A and miR-3682 were both knocked down in HepG2 and Huh7 cells, OD values and the number of cell colonies were rescued compared with the inhibitor group (Fig. 4a–c, P < 0.01). Similarly, the numbers of migratory and invasive cells were notably decreased in miR-3682 inhibitor transfected HCC cells compared with NC group, while the transfection of si- ADRA1A produced the opposite effect (Fig. 4d and e). The same result was obtained that si- ADRA1A co-transfection reversed the restraining effects of miR-3682 inhibitor on mobility of HCC cells. Therefore, these results indicated that miR-3682 promoted malignant phenotype of HCC cells by targeting ADRA1A.
MiR-3682 promoted biological behaviors of HCC cells by targeting ADRA1A. (A, B) The proliferation of HepG2 (A) and Huh7 (B) cells transfected with si-ADRA1A and/or miR-3682 inhibitor/NC was measured by CCK-8 assays. (C) Colony formation assay of HCC cells transfected with si-ADRA1A and/or miR-3682 inhibitor/NC. (D, E) The migration and invasion of HepG2 (D) and Huh7 (E) cells transfected with si-ADRA1A and/or miR-3682 inhibitor/NC was measured by transwell assays, magnification × 200. The data are exhibited as mean±SD with triplicates. ⁎⁎P < 0.01 vs. inhibitor NC group. ##P < 0.01 vs. inhibitor group
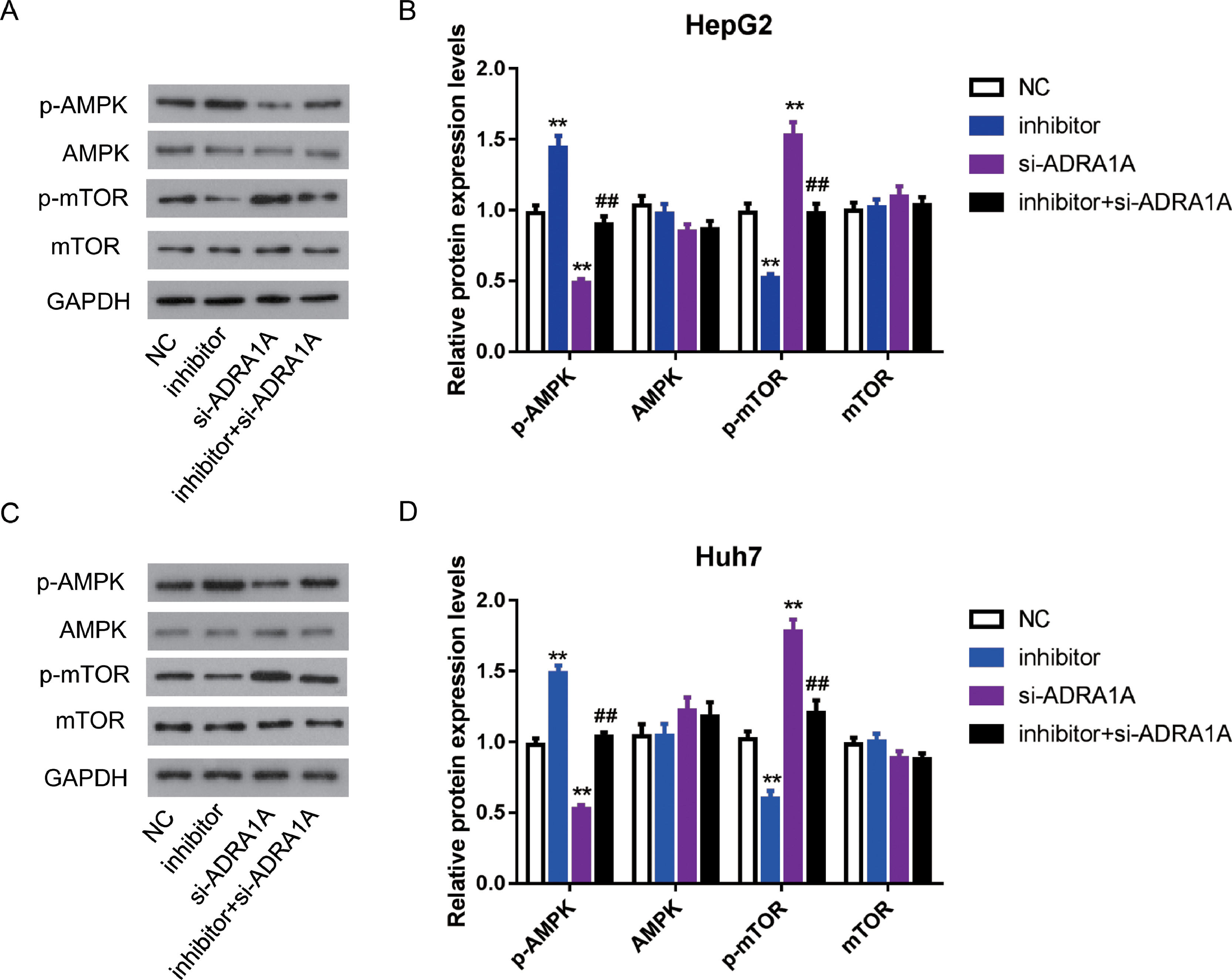
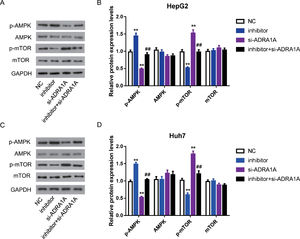
As mentioned above, AMPK signaling pathway was enriched by subjecting 67 genes to DAVID database. Many studies reported that AMPK signaling pathway was involved in various cancers, including HCC [13], so we further investigated whether miR-3682 facilitated malignant phenotype of HCC cells by regulating AMPK signaling pathway. As AMPK is a central regulator of AMPK signaling pathway, meanwhile, mTOR is a key downstream target and negatively regulated by AMPK signaling pathway [14], we measured the expression of AMPK, phosphorylated AMPK (p-AMPK), mTOR and phosphorylated mTOR (p-mTOR) in miR-3682 inhibitor and/or si- ADRA1A transfected HCC cells. As shown in Fig. 5A–D, the level of p-AMPK were notably elevated in miR-3682 inhibitor transfected cells compared to the NC group, while knockdown of ADRA1A caused a reduction of p-AMPK. And loss of ADRA1A reversed the augmentation of p-AMPK level induced by silencing miR-3682 (Fig. 5a–d, P < 0.01). Conversely, inhibition of miR-3682 reduced the phosphorylation of mTOR compared with the NC group, which was increased following the knockdown of ADRA1A, while the reduction were rescued when si- ADRA1A was co-transfected with miR-3682 inhibitor (Fig. 5a–d, P < 0.01). These data supports that miR-3682 inactivates AMPK signaling pathway by suppressing ADRA1A.
MiR-3682 inactivated AMPK signaling pathway via targeting ADRA1A. (A) When HepG2 cells were transfected with si-ADRA1A and/or miR-3682 inhibitor/NC, the expression levels of AMPK, phosphorylated AMPK (p-AMPK), mTOR and phosphorylated mTOR (p-mTOR) were measured by western blot. (B) Quantification of Fig. (A). (C) When Huh7 cells were transfected with si-ADRA1A and/or miR-3682 inhibitor/NC, the expression levels of AMPK, p-AMPK, mTOR and p-mTOR were measured by western blot. (D) Quantification of Fig. (C). The data are exhibited as mean±SD with triplicates. ⁎⁎P < 0.01 vs. inhibitor NC group. ##P < 0.01 vs. inhibitor group
In our study, we found that miR-3682 was up-regulated in HCC tissues and cells and associated with the death of HCC patients. The expression of miR-3682 was a negative and independent prognostic factor of HCC patients. Furthermore, the inhibition of miR-3682 suppressed HCC cell viability, migration and invasion. Consistently, in a previous study, the same results were obtained that miR-3682 contributed to the biological behaviors of colon adenocarcinoma cells [12]. This previous study establishes a basis for our results that miR-3682 plays an oncogenetic role in HCC and may be a potential negative prognostic indicator for HCC.
α1A- Adrenergic receptor (ADRA1A), a subtype of α1- Adrenergic receptor (AR), belongs to the family of G protein-coupled receptors (GPCR) [15]. ARs, stimulated by endogenous catecholamines norepinephrine (NE) and epinephrine, can activate the sympathetic nerves, which is critical to stress response [16]. Previous epidemiological studies reveal that drugs lowering NE level significantly affect the incidence of several cancers, such as prostate, breast and lung cancers [17]. Further, multiple studies have revealed that α1-ARs are involved with the progression of many cancers. α1-ARs are comprised of three members, including ADRA1A, ADRA1B and ADRA1D, among which, ADRA1A is ubiquitously expressed on cell surface and inside the cells [18]. In our study, ADRA1A was firstly identified as a direct target of miR-3682. ADRA1A was down-regulated in HCC and independently associated with a favorable prognosis of HCC patients. Besides, ADRA1A restrained HCC cell viability and mobility. Thus, our study suggested a tumor suppressive role of ADRA1A in HCC. In line of our investigation, bioinformatics analysis in two studies demonstrated that ADRA1A was down-regulated in HCC and positively correlated with the prognosis of HCC patients [19,20]. Besides, a systematic analysis revealed that ADRA1A was an alternative splicing gene, positively associated with the prognosis of renal clear cell carcinoma [21]. However, adverse effects of ADRA1A on many other cancers were obtained. For example, ADRA1A was down-regulated in gastric cancer and negatively associated with the outcome of gastric cancer patients [22]. In hysterocarcinoma, serum ADRA1A was highly expressed and negatively correlated with the prognosis of hysterocarcinoma patients. And serum ADRA1A was associated with the FIGO staging and the presence of lymph node metastasis [23]. These studies implicated that ADRA1A may exert different roles in different cancers.
AMPK, a serine/threonine kinase, is highly conserved across most eukaryotes [24]. AMPK signaling is critical to the regulation of cell homeostasis as a cellular metabolic sensor [24]. Multiple downstream molecules of AMPK signaling pathway, such as TSC2, an anti-oncogene, participate in the regulation of cell cycle and proliferation [13]. Beside, AMPK signaling participates in the regulation of several cancer-related pathways, including rapamycin (mTOR) signaling pathway, which is critical to cell growth, survival and apoptosis [13,25]. Importantly, lines of evidence have demonstrated that AMPK is associated with tumorigenesis and development of various cancers, including HCC [26–28]. Zheng et al. reported that the low level of p-AMPK (Thr172) was involved in the poor prognosis and malignancy of HCC and the stimulation of AMPK signaling by metformin significantly hindered HCC cell growth [29]. Consistently, several investigations about the analysis of phosphorylation of AMPK (Thr172) in HCC patients identified a low level of p-AMPK in HCC patients [30,31,29]. And the higher expression of p-AMPK was associated with a lower risk of HCC reoccurrence and a longer overall survival time of HCC patients [30,31,29]. Furthermore, dysfunction of AMPK signaling plays an important role in HCC cell viability, metastasis and survival. The activation of AMPK could hinder HCC cell proliferation by regulating cell cycle-related genes, such as P27, P21 and cyclin D1 [32–34]. Besides, AMPK signaling blocked the synthesis of protein and inhibited HCC cell proliferation [35]. Of note, several studies demonstrated that AMPK signaling pathway blocked the downstream pathway of mTOR by inhibiting the phosphorylation of mTOR and further restrained HCC cell proliferation and mobility [36,37]. Moreover, several miRNAs, such as miR-519b and miR-23a, were reported to be involved in the regulation of AMPK signaling [38,39]. Similarly, in the current study, miR-3682 inhibited AMPK signaling by targeting ADRA1A, which can be classified as that ADRA1A is an upstream protein of AMPK and directly regulates the activation of AMPK in the signaling. Combined with the aforementioned investigations, our results implicates that miR-3682 inactivates AMPK signaling by inhibiting ADRA1A to facilitate HCC cell proliferation and mobility.
5ConclusionTaken together, for the first time, our study suggests that miR-3682 expression is associated with the death and poor prognosis of HCC patients, and miR-3682 exerted an oncogenetic role in HCC. Furthermore, ADRA1A, identified as a novel target gene of miR-3682, exerts a tumor suppressive effect on HCC. Besides, we clarify that miR-3682 promotes HCC cell malignant phenotype through inhibiting AMPK signaling via targeting ADRA1A. Altogether, our investigation implicates that miR-3682 is an oncogenic miRNA and has the potential to be a therapeutic biomarker and prognostic indicator of HCC.
CRediT authorship contribution statementWenyue Zhao: Formal analysis, Writing – original draft, Writing – review & editing. Xueping Liu: Visualization, Conceptualization, Project administration, Writing – original draft, Writing – review & editing.