Background. Heme oxygenase-1 (HMOX1) and bilirubin UDP-glucuronosyltransferase (UGT1A1), both enzymes involved in bilirubin homeostasis, play an important role inoxidative stress defense.
Objective. To assess the effect of promotervariations of HMOX1 and UGT1A1 genes on the progression of fibrosis in patients chronically infected with the hepatitis C virus (HCV).
Material and methods. The study was performed on146 chronic HCV infection patients, plus 146 age- and sex-matched healthy subjects. The (GT)n and (TA)n dinucleotide variations in HMOX1 and UGT1A1 gene promoters, respectively, were determined by fragment analysis in all subjects.
Results. No differences were found in the frequencies of each particular allele of both genes, between HCV patients and a control group (p > 0.05). Furthermore, no association was detected (p > 0.05) between either the HMOX1 or the UGT1A1 promoter variants and the individual histological stages of liver disease in the HCV positive patients. Finally, no differences in the HMOX1 and UGT1A1 genotype prevalence rates were found between pre-cirrhotic and cirrhotic patients (p > 0.05).
Conclusion. Based on our data, microsatellite variations in the HMOX1 and UGT1A1 genes are not likely to protect from progression of liver disease in patients with chronic hepatitis C.
During the past decades, the heme catabolic pathway has been demonstrated as making an important contribution to the protection against oxidative stress and inflammation. Heme oxygenase (HMOX), the first and rate-limiting enzyme in the heme cata-bolism, leads to equimolar production of carbon monoxide, free iron, and biliverdin (which in turn is reduced to bilirubin, the major intravascular bile pigment). Two isoenzymes of HMOX (HMOX1, OMIM *141250, and HMOX2, OMIM *141251) exist in the human body.1 HMOX1 is highly inducible by many stressful factors, including oxidative stress and inflammatory stimuli; whereas, HMOX2 is constitutively expressed. Hepatic bilirubin UDP-glu-curonosyl transferase (UGT1A1, OMIM *191740), the other important enzyme in the heme catabolic pathway, is responsible for the conjugation of biliru-bin with glucuronic acid; accordingly, facilitating thus its elimination into the biliary system. Congenital reduction in bilirubin glucuronidation (to approximately 30%) results in mild, chronic, fluctuating unconjugated hyperbilirubinemia (Gilbert’s syndrome).2
A number of recent studies have shown that specific promoter variations in both the HMOX1 and UGT1A1 genes are associated with various pathological conditions (for review see ref. 3). The HMOX1 gene promoter contains a highly polymorphic dinucleotide GT repeats (ranging from 11 to 42), which are responsible for the regulation of HMOX1 gene transcription.4 Subjects with the less active long (L) allele (GT ≥ 29) have been shown to exhibit an increased risk of oxidative stress-mediated conditions including cardiovascular diseases, as well as certain forms of cancer.3UGT1A1 also has a highly polymorphic promoter region with (TA)n repeat variations, modulating the UGT1A1 transcriptional activity with clinically important consequences similar to those of the HMOX1 promoter gene variants.3
Among other factors, oxidative stress makes an important contribution to the pathogenesis of liver fibrosis in chronic HCV infection.5 It has been reported by Mori, et al., that cells transfected with cDNA of the HCV core antigen exhibited elevated levels of reactive oxygen species (ROS).6 Therefore it is surprising, that recent data suggest that HMOX1 might also play a role in the pathogenesis of chronic HCV infection. Abdalla, et al., recently described a decrease of HMOX1 protein and mRNA in liver samples from patients infected with HCV.7 Such down regulation of HMOX1 is likely caused by the HCV core protein, rather than by the non-structural HCV proteins.8 In contrast, Ghaziani, et al. found upregulation of HMOX1 in the human hepatoma cells expressing HCV proteins from the core region, up to the aminotermi-nal domain of the NS3 region of the HCV genome.9 The importance of HMOX1 on HCV replication was proven by Zhu, et al., who transfec-ted human hepatoma cells harboring HCV replicons with a human HMOX1 gene.10 HCV replication was inhibited in cells overexpressing HMOX1; and this was reversed with siRNA-media-ted HMOX1 knockdown. Moreover, HMOX1 induction with hemin also significantly decreased HCV replication. These findings suggest that the pharmacological activation of HMOX1 might provide an adjuvant benefit to the standard antiviral treatment options and/or protect from hepatocellu-lar injury in chronic HCV infection. This concept is supported by the recent investigation of Lehman, et al., who demonstrated that HMOX1 induction significantly inhibited HCV replication by increasing interferon response in vitro.11 On the other hand, Bonkovsky, et al., in their clinical study did not find any association of the variations of HMOX1 gene promoter and responses to the first phase of antiviral therapy, or to the likelihood of developing outcomes in a HALT-C trial.12
Based on this data, we focused on assessing the role of HMOX1 and UGT1A1 promoter variations in liver disease progression in chronically HCV-infec-ted patients.
Material and MethodsPatientsThe study was performed on 146 patients diagnosed with chronic HCV infection, who were followed at the Department of Internal Medicine of 1st. Faculty of Medicine, Charles University in Prague and Central Military Hospital, Prague, Czech Republic.
Chronic HCV infection was defined as the HCV RNA positivity by RT-PCR in the serum for at least 6 months, and in whom other possible causes of liver disease were excluded (including HBV or HIV coinfections). A liver biopsy was performed on 112 patients by the aspiration technique. The transcuta-neous approach was used in the majority of cases, while transjugular biopsies were performed on 23 patients. Tissue samples were evaluated according to the Ishak scoring system (as has been described previously).13 All samples were evaluated by a single experienced pathologist (P.H. Central Military Hospital Prague). In these patients, complete data on the grading and staging of the liver disease was available. A liver biopsy was refused by 19 patients; however, none of these patients showed clinical signs of liver cirrhosis. Clinically evident liver cirrhosis was found in 15 patients (signs of portal hypertension, hypersplenism, or a history of liver decompensation). Bioptically-proven liver cirrhosis (stage 6) was detected in 10 patients; together forming the group of 25 liver cirrhosis patients (Figure 1). The control group consisted of 146 age- and sex-matched healthy blood donors.
The study’s protocol conformed to all of the ethical guidelines in the 1975 Declaration of Helsinki, as well as being approved by the Ethics Committee of the respective institutions.
The study was registered on www.clinicaltrials.gov (ClinicalTrials.gov Identifier NCT00842205).
MethodsThe (GT)n variations in HMOX1 (dbSNP rs1805173) and (TA)n variations in the UGT1A1 (dbSNP rs81753472) gene promoters were determined by fragment analysis as described previously.14 In brief, corresponding DNA fragments were amplified by a duplex polymerase chain reaction (PCR), using the following primers:
- •
HMOX1
- °
Forward: 5’ -ctgcagcttctcagatttcc - 3’.
- °
Reverse: 5’ - acaaagtctggccataggac - 3’.
- •
UGT1A1
- °
Forward: 5’ -gaacttggtgtatcgattggtttttgc - 3’.
- °
Reverse: 5’ - catccactgggatcaacagtatcttcc - 3’.
The reverse primers were labeled at the 5’ end with WellRED fluorescent dyes (Beckman Coulter, Fullerton, CA, USA). The resulting PCR products were separated on a CEQ 8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA, USA). Based on the number of (GT)n repeats, HMOX1 alleles were classified into short (S, n < 24), medium (M, n = 24–28) and long (L, n ≥ 29) subgroups.15,16
The distribution of genotypes in the control subjects was within the Hardy-Weinberg equilibrium for both studied loci.
HCV RNA was detected in the plasma by use of the Cobas Ampliprep/TaqMan system (Roche, Lower limit of Detection, LLD = 15 IU/mL).
Statistical analysesThe analyses comparing the frequency rates of different alleles as markers tophenotype parameters were based on the Fisher exact test. All analyses were performed with alpha set to 0.05.
ResultsHMOX1 promoter variants in HCV-infected patientsThe frequencies of S, M, and L alleles of the HMOX1 gene promoter did not differ between the HCV positive patients and control subjects (p > 0.05, data not shown). Analogously, no differences were found between the cases and controls for the individual HMOX1 genotypes (p = 0.40, data not shown).
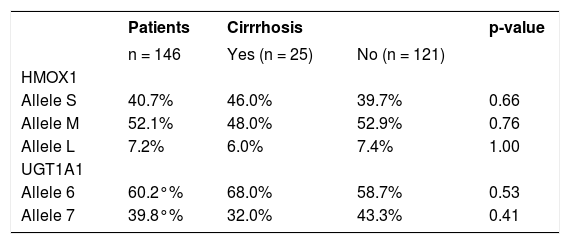
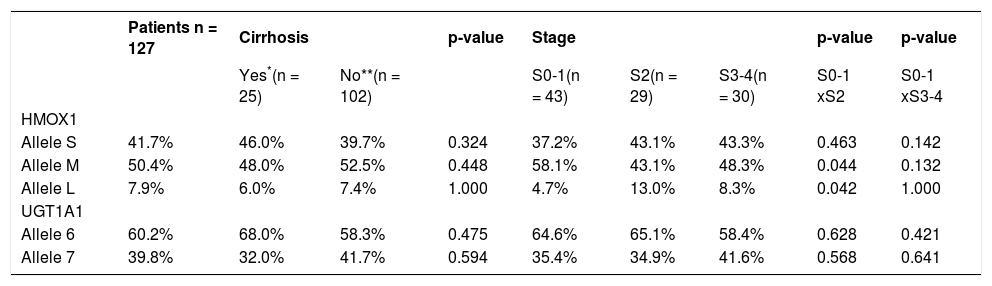
In addition, no differences were also detected in the frequencies of the individual alleles between HCV patients without liver cirrhosis and those with overt liver cirrhosis (Table 1, p > 0.05 for all comparisons). This observation was confirmed in the detailed analysis of the possible role of HMOX1 pro-moter gene variants on disease progression in HCV-infected patients who had undergone a liver biopsy. For this purpose, we grouped non-cirrhotic patients according to the staging into three subgroups: S 0–1 (n = 43), S 2 (n = 29) and S 3–4 (n = 30). There were no significant associations between the frequency of individual HMOX1 alleles and the stage of the liver lesion (Table 2, p > 0.05 for all comparisons). As expected, patients with either a more advanced liver fibrosis, or cirrhosis, were significantly older than were patients with lower stages (p < 0.005, data not shown).
Frequency of HMOX1 and UGT1A1 alleles in HCV patients with and without liver cirrhosis
| Patients | Cirrrhosis | p-value | ||
|---|---|---|---|---|
| n = 146 | Yes (n = 25) | No (n = 121) | ||
| HMOX1 | ||||
| Allele S | 40.7% | 46.0% | 39.7% | 0.66 |
| Allele M | 52.1% | 48.0% | 52.9% | 0.76 |
| Allele L | 7.2% | 6.0% | 7.4% | 1.00 |
| UGT1A1 | ||||
| Allele 6 | 60.2°% | 68.0% | 58.7% | 0.53 |
| Allele 7 | 39.8°% | 32.0% | 43.3% | 0.41 |
Data are given as percentage of a given allele.
Association between HMOX1 and UGT1A1 alleles, and the severity of liver disease.
| Patients n = 127 | Cirrhosis | p-value | Stage | p-value | p-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Yes*(n = 25) | No**(n = 102) | S0-1(n = 43) | S2(n = 29) | S3-4(n = 30) | S0-1 xS2 | S0-1 xS3-4 | |||
| HMOX1 | |||||||||
| Allele S | 41.7% | 46.0% | 39.7% | 0.324 | 37.2% | 43.1% | 43.3% | 0.463 | 0.142 |
| Allele M | 50.4% | 48.0% | 52.5% | 0.448 | 58.1% | 43.1% | 48.3% | 0.044 | 0.132 |
| Allele L | 7.9% | 6.0% | 7.4% | 1.000 | 4.7% | 13.0% | 8.3% | 0.042 | 1.000 |
| UGT1A1 | |||||||||
| Allele 6 | 60.2% | 68.0% | 58.3% | 0.475 | 64.6% | 65.1% | 58.4% | 0.628 | 0.421 |
| Allele 7 | 39.8% | 32.0% | 41.7% | 0.594 | 35.4% | 34.9% | 41.6% | 0.568 | 0.641 |
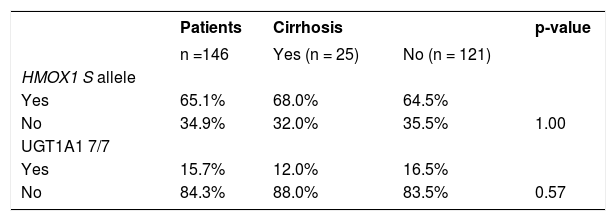
As the S allele of the HMOX1 gene seems to be associated with a lower incidence of oxidative stress-related diseases, we have analyzed in detail the frequency of this particular allele (genotypes SS/SL/ SM) in the HCV infected patients with liver cirrhosis. Similarly, no difference was detected from this analysis (Table 3, p = 1.00).
UGT1A1 promoter variants in HCV-infected patientsIn a comparable manner, non-significant findings were also detected for the UGT1A1 promoter gene variants. The frequencies of alleles 6 and 7 of the UGT1A1 gene promoter did not differ between HCV positive patients and the healthy controls (data not shown, p > 0.05).
The frequencies of the UGT1A1 alleles in HCV positive patients are shown in table 1. No differences in the frequencies of alleles 6 and 7 of the UGT1A1 gene were found between cirrhotic and non-cirrhotic HCV patients (Table 1, p = 0.53). In addition, no differences were also found in the more detailed analysis, when the non-cirrhotic patients were grouped according to their fibrosis stage upon liver biopsy (Table 2).
As the UGT1A1 7/7 genotype has been reported to be associated with a lower incidence of oxidative stress-related diseases, we have analyzed the occurrence of this particular genotype in patients with or without liver cirrhosis compared to genotypes 6/7 and 6/6 (Table 3). However, in this analysis, no significant differences were found.
HMOX1 and UGT1A1 promoter variants and HCV RNA viral loadIn the final analysis, we focused on the possible protective role of both genetic variants on the HCV RNA viral load. The baseline viral load was considered low if the HCV RNA level at baseline (immediately before treatment initiation) was < 800,000 IU/ mL. Nevertheless, no significant difference in the viral load was found in relationship to the presence of the HMOX1 allele S or the UGT1A1 7/7 genotype (Table 4).
DiscussionThe physiological role of HMOX in heme catabo-lism has been recognized for almost four decades.17 However, its role in mediating protection from inflammation and oxidative stress via the production of bi-lirubin was confirmed only in 1987.18 Several investigators have proven that the induction of HMOX1 can prevent or diminish the severity of liver injury (at least in animal models).19 To date, the precise mechanisms of the protective effects of the HMOX1 have yet to be elucidated. These may involve the degradation of prooxidants, such as heme; the increased generation of antioxidants such as ferritin, biliverdin, bilirubin even carbon monoxide (a bioactive gaseous molecule).
The mechanisms of liver injury in chronic HCV infection are not completely understood. It is believed that several mechanisms might be involved; including immune cell mediated injury, apoptosis, as well as oxidative stress. Pianko, et al. found significantly greater apoptosis in patients with chronic HCV infection, compared with the healthy con-trols.20 Apoptosis is a likely mechanism of liver cell injury in chronic hepatitis C patients with persistently normal ALT. Ming-Ju, et al., recently reported recently increased levels of proinflammatory and pro-fibrotic mediators in the HuH7 cell line; expressing the HCV E2 protein.21 This study indicates that the E2 protein is involved in the pathogenesis of HCV mediated fibrosis by the up-regulation of collagen α and oxidative stress, which is Janus kinase (JAK1) as well as JAK2 pathway-related. Lin and coworkers provided evidence that HCV enhances the progression of hepatic fibrosis through the generation of ROS.22 The other possible factors contributing to liver inflammation in chronic HCV are viral proteins such as the HCV core antigen.23 Ghaziani, et al. found the upregulation of HMOX1 in a cell line expressing several HCV proteins, including core protein.9 HMOX1 in human hepatoma cells may be induced by iron exposure. Furthermore, it has been proven that iron inhibits the expresion of HCV in vi-tro.24 This fact might be of particular interest, as iron 12 overload is negatively associated with the stage of liver disease, and treatment efficacy in HCV infection; and, iron itself is involved in the pathoge-nesis of liver inflammation.25
The presence of the short allele of the highly polymorphic HMOX1 gene promoter was reported to be associated with higher HMOX1 inducibility, leading to increased cytoprotection. Indeed, higher basal HMOX1 expression, as well as stronger inducibility in human endothelial cells carrying the S allele, exposed to oxidative and inflammatory stimuli, has recently been described.26 Based on this data, we have hypothesized that the genetic variant in the HMOX1 gene promoter might affect the response to chronic HCV infection, as well as the progression of liver disease in chronically HCV infected patients. However, our data, however, do not support this hypothesis. We were not able to prove any hepatoprotective effect of the S allele HMOX1 carrier status, either on the HCV infection risk or on liver disease progression.
Similar findings were also obtained also for UGT1A1, another important heme catabolic gene. In contrast to other oxidative stress-mediated diseases, such as atherosclerosis,3 no associations were found for major UGT1A1 gene promoter variations; either in HCV infection risk or liver disease progression. In addition, we were not able to find any association of the biologically relevant variants of both UGT1A1, as well as the HMOX1 genes and hepatitis C viral load.
The lack of any such associations in our study may have been due to several factors. Liver fibrosis in HCV infection is a very complex process, in which many genetically determined factors as well as viro-logical or environmental factors, can play a role. Important factors possibly contributing to the pathogenesis of liver injury in HCV patients such as: the patients age at the time of infection, alcohol consumption, body mass index, insulin resistance, past HBV infection, or hepatic iron content were not available; therefor, these could not be controlled for. In general, taken together, these factors are likely to contribute more significantly to the pathogenesis of liver injury in HCV infection compared to the possible protective role of HMOX1/UGT1A1 promoter gene variants.
Nevertheless, our findings are in accord with a recent clinical study by Bonkovsky, et al., who also found no association between HMOX1 promoter gene variants and the response to HCV infection therapy in chronically infected patients.12
ConclusionOur study in human patients with chronic HCV infection did not prove any association between promoter variants of the HMOX1 and UGT1A1 genes and the risk of disease progression.
Abbreviations- •
CI: Confidence interval.
- •
HBV: Hepatitis B virus
- •
HCV: Hepatitis C virus.
- •
HCV RNA: Hepatitis C virus ribonucleic acid.
- •
HIV: Human inmmunodeficiency virus.
- •
HMOX: Heme oxygenase.
- •
LLD: Lower limit of detection of polymerase chain reaction.
- •
LVL: Low viral load.
- •
OR: Odds ratio.
- •
PCR: Polymerase chain reaction.
- •
ROS: Reactive oxygen species.
- •
SNP: Single nucleotide polymorphism.
- •
SVR: Sustained virlogical response.
- •
UGT1A1: Bilirubin UDP-glucuronosyltransferase.