Introduction and aim. Hepatocellular carcinoma (HCC) is a lethal malignancy, but the molecular mechanisms of hepatocarcinogenesis remain undefined. The present study aims to investigate the relationship between polymorphisms of the hepatic lipase (HL) gene promoters and risk of HCC.
Material and methods. Totally, 279 HCC patients and 200 healthy individuals were enrolled. Polymerase chain reaction-restriction fragment length polymorphisms (PCR-RFLP) was used to analyze the genotypes of HL gene. Logistic regression analysis was conducted to identify risk factors of HCC.
Results. There was significant difference in the distribution of smoking history, drinking history, and family history of subjects between the case and control groups (all p < 0.05). Difference in the -250G/A (p = 0.011; OR = 1.61; 95%CI: 1.11-2.34) and -514C/T (p = 0.007; OR = 1.65; 95%CI: 1.14-2.38) genotypes and allele frequencies between two groups was significant. A higher risk of HCC was identified in those with polymorphisms in the -250G/A (p = 0.007; OR = 1.45; 95%CI: 1.11-1.89) and -514C/T (p = 0.003; OR = 1.51; 95%CI: 1.15-2.00). Polymorphisms at -250G/A (GA + AA) (p = 0.025; OR = 1.55; 95%CI: 1.06-2.28), -514C/T (CT + TT) (p = 0.021; OR = 1.57; 95%CI: 1.07-2.29), smoking history (p = 0.017; OR = 1.70; 95%CI: 1.10-2.63) and drinking history (p = 0.003; OR = 2.04; 95%CI: 1.27-3.27) were significantly related to the risk of HCC (all p < 0.05).
Conclusion. The results obtained from this study indicated that polymorphisms of -250G/A and -514C/T in HL gene promoters were associated with the risk of HCC.
Hepatocellular carcinoma (HCC), one of the most common malignancies, ranks the fifth in cancer incidence and the third in cancer mortality around the world; over 1.2 million people die from it yearly, especially in Asia and Southern Africa.1,2 It is worth noting that the HCC incidence and mortality is the third among all malignancies in China.3 At present, surgical resection and liver transplantation are the two main treatments for early-stage HCC, but the HCC prognosis remains poor and unstable due to tumor invasion, metastasis and recidivism, and chemotherapy resistance.1 Risk factors for HCC generally include hepatitis B and C virus infection, obesity, dietary carcinogens and iron overload.4 Cigarette smoking has been suggested as a causal factor of HCC progression, as well as a prognostic risk factor for HCC patient survival.5,6 Based on the study of Cleary, et al., genetic alterations are crucial factors in the occurrence and development of HCC, since they result in disorders of cellular pathways.7 However, not all individuals exposed to risk factors develop HCC because of individual differences, and studies on susceptibility of HCC are beneficial for illuminating the molecular basis of individual differences.8 Multiple studies have exerted their efforts to genetic correlations between specific driver gene polymorphisms and the risk of HCC.9,10
As the most common genetic variations exist in human body,1 single nucleotide polymorphisms (SNPs) are associated with disease susceptibility and treatment outcomes, and provide powerful tools for a variety of medical genetic studies.11,12 Hepatic lipase (HL), synthesized and secreted by hepatic parenchymal cells, is a member of aliphatic families, which form a complex by the action with heparan sulfate proteoglycans (HSPG).13 HL promoter has four common SNPs linked to inheritance, including -250G/A, -514C/T, -763A/G, and -710T/C, which affect the metabolism of plasma lipoproteins; moreover, there exists association between promoter polymorphisms of HL gene, HL activity, and plasma lipoprotein.14 HCC is considered as a multifactorial disease and gene polymorphisms is regarded as an important contributor to hepatocarcinogenesis, indicating a high degree of genetic heterogeneity might be exhibited in HCC patients.8 At present, studies on the HL gene polymorphisms and HCC risk has been not found yet. In this study, polymerase chain reaction restriction fragment length polymorphisms (PCR-RFLP) was used to detect the genotypes of -250G/A, -514C/T, -763A/G, and -710T/C in patients with HCC in order to clarify the role of HL gene promoter polymorphisms in the occurrence of HCC.
Material and MethodsEthic statementThis study was conducted with the approval of the Ethics Committee of Yantai Yuhuangding Hospital. Informed consent and required documentation was obtained from each patient and respective guardians prior to the study.
Study subjectsFrom September 2013 to September 2015, 279 patients with HCC received clinical and pathological diagnosis in Yantai Yuhuangding Hospital were collected as the case group. All the included patients were new cases, with secondary cases and recurrent cases of HCC excluded. Among 279 patients, there were 143 male patients (aging 30-75 years old) and 136 female patients (aging 51.17 ± 10.30 years old). There were 90 smoking patients, 189 nonsmoking patients; 96 drinking patients, 183 non-drinking patients and 35 patients with family history. During the same period, 200 healthy individuals who came to Yantai Yuhuangding Hospital for physical examination were enrolled as the control group. Among the healthy controls, there were 103 male patients (aging 30-75 years old) and 97 female patients (aging 52.34 ± 10.70 years old), and there were 43 smoking patients, 157 non-smoking patients; 37 drinking patients, 163 non-drinking patients and 11 patients with family history. In both of the case and control groups, individuals were excluded with infection of hepatitis B, hepatitis C virus infection, or other liver diseases, including alcoholic liver disease, metabolic liver disease. People of the Han nationality without blood relationship were recruited, whose routine tests were normal.
Questionnaire surveyIn this study, detailed demographic data and related exposure factors of patients in the case group were obtained from records of Yantai Yuhuangding Hospital, and main contents included age, gender, smoking history, drinking history, family history. Informed consent was obtained from each patient. The self-made questionnaires (Table 1) about demographic data and related exposure factors were used for people in the control group, including age, gender, smoking history, drinking history, family history. Smoking history standard was defined as six months or more of continuous or cumulative smoking in lifetime, and drinking history was defined as drinking at least once a week for six months or more. After obtaining the informed consent, professionally trained investigators were dispatched to conduct a detailed inquiry of the subjects face-to-face, and results were filled in the questionnaire according to the facts.
Health Questionnaire.
| Number of vour hospital: |
| Section 1 Personal information |
| 1 Name: |
| 2. Age: |
| 3. Sex: male □ female □ |
| 4. Nationality: Han nationality □ other nationalities □ |
| 5. Marital status: single □ married □ re-marriage □ divorced □ widowed □ |
| 6. Occupation: worker □ peasant □ office clerk □ Medical and Health staff □ |
| Self-employed administrator □ others □| |
| 7. Native place: |
| 8. Birthplace: |
| 9. Address: Phone: |
| 10. Smoking history: □ Yes □ No (Turn to the 13th question with the answer No) |
| 11. Duration of cigarette smoking: |
| 12. Smoking frequency: pack/week |
| 13. Drinking history: Yes No (Turn to the 16th question with the answer No) |
| 14. Drinking age: year |
| 15. Drinking frequency: □ once a week □ twice to forth a week □ fifth to sixth a week |
| □ once a day □ twice or more than twice a day |
| 16. Family history: □ Yes □ No |
| 17. Height: |
| 18. Weight: |
| Section 2 Results of physical and laboratory examinations |
| 1. Blood type: |
| 2. HBV test |
| Hepatitis B surface antigen (HBsAg): Hepatitis B e antibody (anti-HBe): |
| Hepatitis B surface antibody (anti-HBs): Hepatitis B core antibody (anti-HBe): |
| Hepatitis B e antigen (HBeAg): HBV-DNA: |
| 3. Liver functions |
| Total bilirubin (TBIL) 3-22 umol/LDirect bilirubin (DBIL) 0-6 umol/L |
| Indirect bilirubin (IBIL) 3-22 umol/LTotal protein (TP) 60-80 g/L |
| Albumin (ALB) 35-55 g/L Globulin (GLO) 25-35 g/L |
| r-Transpeptidase (GGT) 0-40 U/LAlanine aminotransferase (ALT) 8-45 U/L |
| Aspartate aminotransferase (AST) 4-40 U/L |
| Alkaline phosphatase (ALP) 40-150 U/L |
| 4. Liver fluke enzyme marker: |
| 5. Anti-KCV: |
| 6. AFP content: 0-10.9 ng/mL |
Five mL of venous blood was collected from subjects with empty stomach in the morning, and anticoagulated with elhylene diamine tetraacetic acid (EDTA) (Shanghai Haling Biological Technology Co. Ltd., Shanghai, China). According to the kit instructions, DNA extraction kit was used to extract the genomic DNA. Nucleic acid-protein detector (Puyang Scientific Instrument Research Institute of Nanjing University, Nanjing, Jiangsu, China) was used for quantitative analysis. The extraction was stored at -20 °C for preservation. Blood genomic DNA extraction kit was purchased from Shanghai Sangon Biotech Co., Ltd. (Shanghai, China).
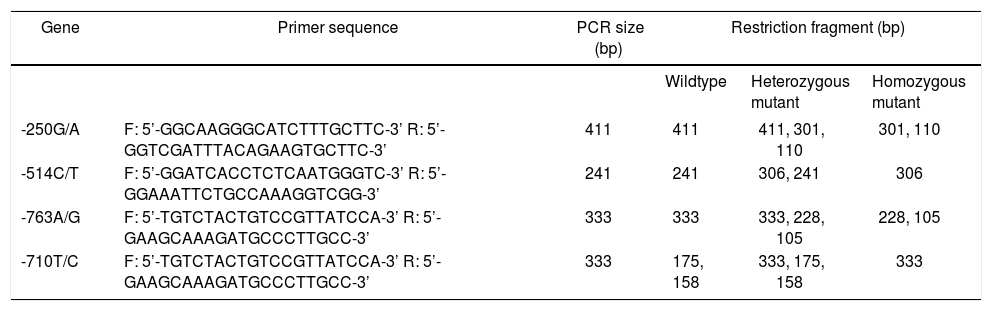
GenotypingPolymorphisms analysis of genes was conducted by PCR-RFLP. The 25 μL of PCR reaction system contains 1 x PCR buffer, 1.5 mmol/L Mg2+, 200 μmol/L dNTP, 200 ng of DNA template, 1 U TaqDNA polymerase (Promega Corporation, Madison, WI, USA) and 200 nmol/L of both forward and reverse primers. PCR reaction conditions: pre-denaturation at 94 °C for 5 min, denaturation at 94 °C for 40 s, annealing at 55 °C for 40 s, extension at 72 °C for 90 s, which ran for 35 cycles, followed by extension at 72 °C for 5 min. PCR amplification was determined by 2% agarose gel electrophoresis containing 0.5 μg/mL ethidium bromide (Beijing RuiDaHengHui Science and Technology Development Co., Ltd, Beijing, China). PCR primers were synthesized by Shanghai Invitrogen Biotech Co., Ltd. (Shanghai, China) (Table 2). Extracted agarose gel was treated with electrophoresis (5 V/cm) for 40 min, and the reaction products were observed under a ultraviolet (UV) light. Then 5 μL of PCR products were added with 10 of endonuclease DRA I, NLA III, SPH I, and AVA II (TaKaRa Biotechnology Dalian Co., Ltd., Dalian, Liaoning, China) respectively, and placed at a constant temperature overnight. Enzyme-digested products were treated with 4% agarose gel electrophoresis and 8% non-denaturing polyacrylamide gel (SNF Floerger, France) electrophoresis and then stained by 0.1% AgNO3 (Bolinda Technology Co., Ltd., Shenzhen, Guangdong, China) to observe the genotypes.
Primer sequences for PCR-RFLP.
| Gene | Primer sequence | PCR size (bp) | Restriction fragment (bp) | ||
|---|---|---|---|---|---|
| Wildtype | Heterozygous mutant | Homozygous mutant | |||
| -250G/A | F: 5’-GGCAAGGGCATCTTTGCTTC-3’ R: 5’-GGTCGATTTACAGAAGTGCTTC-3’ | 411 | 411 | 411, 301, 110 | 301, 110 |
| -514C/T | F: 5’-GGATCACCTCTCAATGGGTC-3’ R: 5’-GGAAATTCTGCCAAAGGTCGG-3’ | 241 | 241 | 306, 241 | 306 |
| -763A/G | F: 5’-TGTCTACTGTCCGTTATCCA-3’ R: 5’-GAAGCAAAGATGCCCTTGCC-3’ | 333 | 333 | 333, 228, 105 | 228, 105 |
| -710T/C | F: 5’-TGTCTACTGTCCGTTATCCA-3’ R: 5’-GAAGCAAAGATGCCCTTGCC-3’ | 333 | 175, 158 | 333, 175, 158 | 333 |
Epidata software 3.02 (The Epidata Association, Odense, Denmark) and SSPS 18.0 software (SPSS Inc., Chicago, IL, USA) were used for data collection and analysis. The measurement data was performed as mean ± standard deviation, and t-test was used. The enumeration data were analyzed by Chi-square test. Chi-squared (χ2) goodness-of-fit test was applied to identify the genotypes of the control group, and to determine whether the genotype distribution is consistent with Hardy-Weinberg equilibrium (HWE). Bonferroni correction was used for multiple comparisons. Multivariate logistic regression was used to calculate odds ratios and 95% confidence intervals (CI) for the estimation to the relative odds of related factors and the association between gene polymorphisms and disease. p < 0.05 represented significant difference.
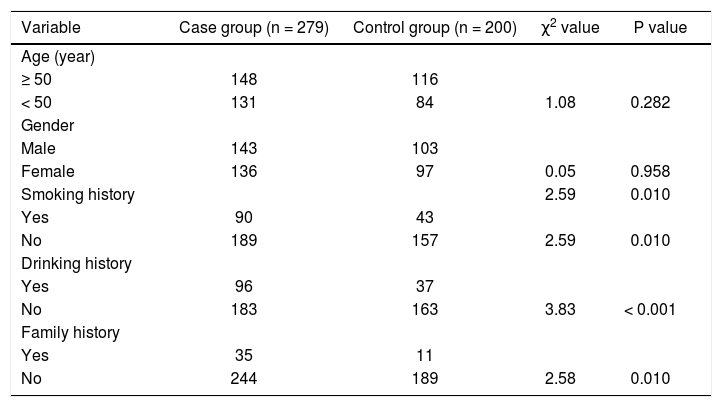
ResultsBaseline characteristics of the case and control groups are comparableA total of 279 patients with HCC were enrolled in this study. Compared with the baseline characteristics of healthy population, there was no significant difference in the distribution of age and gender between the two groups (p > 0.05). Compared with the control group, there was significant difference in the distribution of smoking history, drinking history, and family history in the case group (p < 0.05) (Table 3).
The baseline characteristics of study subjects between the case and the control groups.
| Variable | Case group (n = 279) | Control group (n = 200) | χ2 value | P value |
|---|---|---|---|---|
| Age (year) | ||||
| ≥ 50 | 148 | 116 | ||
| < 50 | 131 | 84 | 1.08 | 0.282 |
| Gender | ||||
| Male | 143 | 103 | ||
| Female | 136 | 97 | 0.05 | 0.958 |
| Smoking history | 2.59 | 0.010 | ||
| Yes | 90 | 43 | ||
| No | 189 | 157 | 2.59 | 0.010 |
| Drinking history | ||||
| Yes | 96 | 37 | ||
| No | 183 | 163 | 3.83 | < 0.001 |
| Family history | ||||
| Yes | 35 | 11 | ||
| No | 244 | 189 | 2.58 | 0.010 |
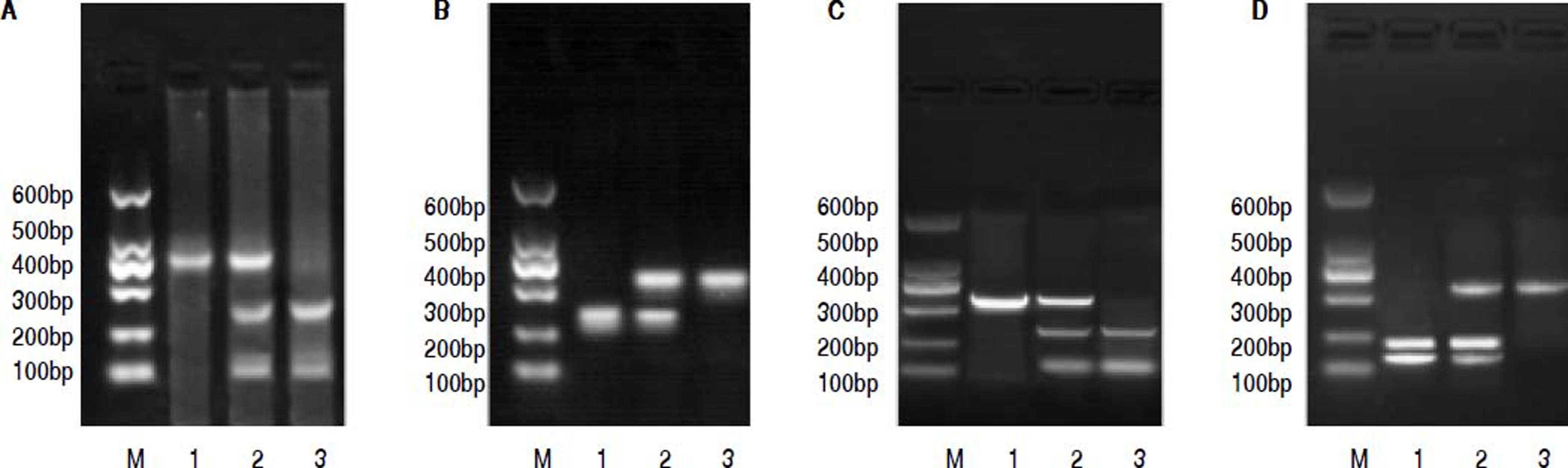
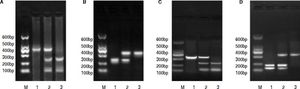
PCR-RFLP was performed to analyze the genotypes of HL gene. The results of -250 G/A were a 411 bp fragment as GG (wildtype), three fragments of 411 bp, 301 bp and 110 bp as GA (heterozygote variant), in addition to two fragments of 301 bp and 110 bp as AA (homozygous mutant) (Figure 1A). The results of -514C/T were a 241 bp fragment as CC type (wildtype), 241 bp and 306 bp fragments as CT (heterozygote variant) as well as a 306 bp fragment as TT (homozygous mutant) (Figure 1B). The result of -763A/G was a 333 bp fragment as AA (wildtype), 333 bp, 228 bp and 105 bp fragments as AG (heterozygote variant), in addition to 228 bp and 105 bp fragments as GG (homozygous mutant) (Figure 1C). The result of -710T/C was 175 bp and 158 bp fragments as TT (wildtype); 333 bp, 175bp and 158 bp fragments as TC (heterozygote variant); a 333 bp fragment as CC (homozygous mutant) (Figure 1D).
The electrophoretogram of PCR-RFLP products. A. -250G/A locus. M: represents marker. 1: represents GG. 2: represents GA. 3: represents AAA. B. -514C/Tlocus. M: represents marker. 1: represents CC. 2: represents CT. 3: represents TT. C. 763A/G locus. M: represents marker. 1: represents AAA: 2: represents AG. 3: represents GG: D. -710T/C locus. M: represents marker. 1: represents TT. 2: represents TC: 3: represents CC. PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism.
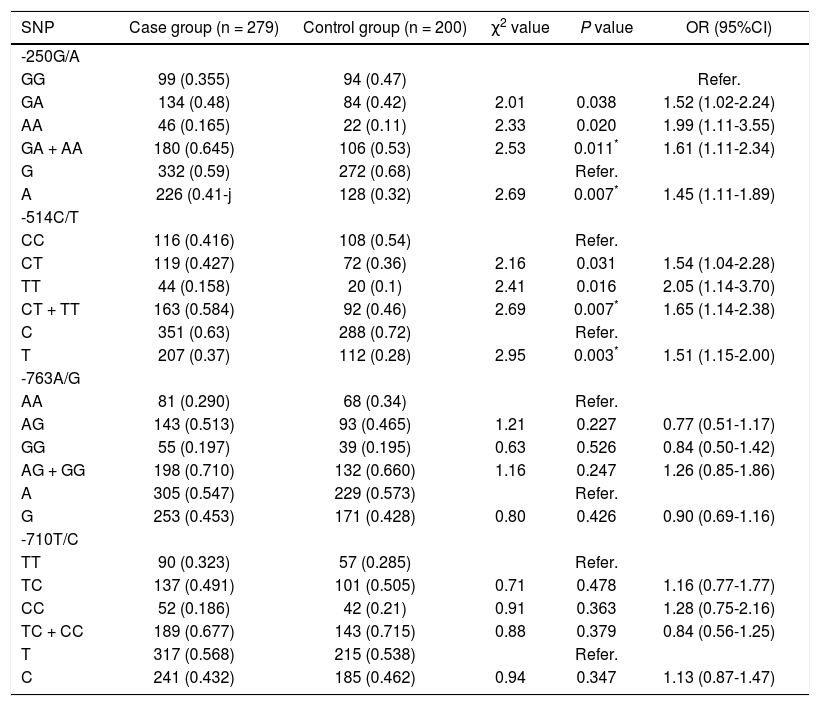
Genotype distribution of polymorphisms and allele frequency of the four promoters of HL as -250G/A, -514C/ T, -763A/G and 710T/C in the control group and the case group are listed in table 4. These promoters reached genetic equilibrium with population representation after the HWE test. Differences in -250G/A and -514C/T genotypes and allele frequencies between the two groups were statistically significant (p < 0.05). No difference in 763A/G and -710T/C genotypes and allele frequencies was found between the two groups (p > 0.05). Our study statistically analyzed the distribution and allele frequencies of the four polymorphisms using the Bonferroni correction method to control the false discovery, and p < 0.0125 (0.05/4) represented significant difference. Therefore, results of Bonferroni correction method showed that there was significant difference in allele frequencies of -250G/A and -514C/T polymorphisms between the case group and the control group (all p < 0.01). As shown in table 4, for -250G/A polymorphisms, compared with patients with GG wildtype, the risk of HCC was significantly increased in patients who carried (GA + AA) heterozygote variant (p = 0.001; OR = 1.61; 95%: 1.11-2.34). For -514C/T polymorphisms, the risk of HCC in patients who carried (CT + TT) heterozygote variant (p = 0.007; OR = 1.65; 95%: 1.04-2.28) was also increased, when compared with patients with CC wildtype. For -250G/A polymorphisms, compared with patients with G allele, the risk of HCC was notably increased in patients who carried A allele (p = 0.007; OR = 1.45; 95%: 1.11-1.89). For -514C/T polymorphisms, the risk of HCC in patients who carried T allele (p = 0.003; OR = 1.51; 95%: 1.15-2.00) was also increased, when compared with patients with C allele. In -763A/G and -710T/C polymorphisms, there was no statistical correlation between the risk of HCC and carriage of heterozygote variant and homozygous variant, compared with patients who carried wildtype (p > 0.05). Results indicate that polymorphisms in the -250G/A and -514C/T increased the risk of HCC.
Genetic correlation between genotype and allele frequency of four promoters of HL gene and the risk of HCC.
| SNP | Case group (n = 279) | Control group (n = 200) | χ2 value | P value | OR (95%CI) |
|---|---|---|---|---|---|
| -250G/A | |||||
| GG | 99 (0.355) | 94 (0.47) | Refer. | ||
| GA | 134 (0.48) | 84 (0.42) | 2.01 | 0.038 | 1.52 (1.02-2.24) |
| AA | 46 (0.165) | 22 (0.11) | 2.33 | 0.020 | 1.99 (1.11-3.55) |
| GA + AA | 180 (0.645) | 106 (0.53) | 2.53 | 0.011* | 1.61 (1.11-2.34) |
| G | 332 (0.59) | 272 (0.68) | Refer. | ||
| A | 226 (0.41-j | 128 (0.32) | 2.69 | 0.007* | 1.45 (1.11-1.89) |
| -514C/T | |||||
| CC | 116 (0.416) | 108 (0.54) | Refer. | ||
| CT | 119 (0.427) | 72 (0.36) | 2.16 | 0.031 | 1.54 (1.04-2.28) |
| TT | 44 (0.158) | 20 (0.1) | 2.41 | 0.016 | 2.05 (1.14-3.70) |
| CT + TT | 163 (0.584) | 92 (0.46) | 2.69 | 0.007* | 1.65 (1.14-2.38) |
| C | 351 (0.63) | 288 (0.72) | Refer. | ||
| T | 207 (0.37) | 112 (0.28) | 2.95 | 0.003* | 1.51 (1.15-2.00) |
| -763A/G | |||||
| AA | 81 (0.290) | 68 (0.34) | Refer. | ||
| AG | 143 (0.513) | 93 (0.465) | 1.21 | 0.227 | 0.77 (0.51-1.17) |
| GG | 55 (0.197) | 39 (0.195) | 0.63 | 0.526 | 0.84 (0.50-1.42) |
| AG + GG | 198 (0.710) | 132 (0.660) | 1.16 | 0.247 | 1.26 (0.85-1.86) |
| A | 305 (0.547) | 229 (0.573) | Refer. | ||
| G | 253 (0.453) | 171 (0.428) | 0.80 | 0.426 | 0.90 (0.69-1.16) |
| -710T/C | |||||
| TT | 90 (0.323) | 57 (0.285) | Refer. | ||
| TC | 137 (0.491) | 101 (0.505) | 0.71 | 0.478 | 1.16 (0.77-1.77) |
| CC | 52 (0.186) | 42 (0.21) | 0.91 | 0.363 | 1.28 (0.75-2.16) |
| TC + CC | 189 (0.677) | 143 (0.715) | 0.88 | 0.379 | 0.84 (0.56-1.25) |
| T | 317 (0.568) | 215 (0.538) | Refer. | ||
| C | 241 (0.432) | 185 (0.462) | 0.94 | 0.347 | 1.13 (0.87-1.47) |
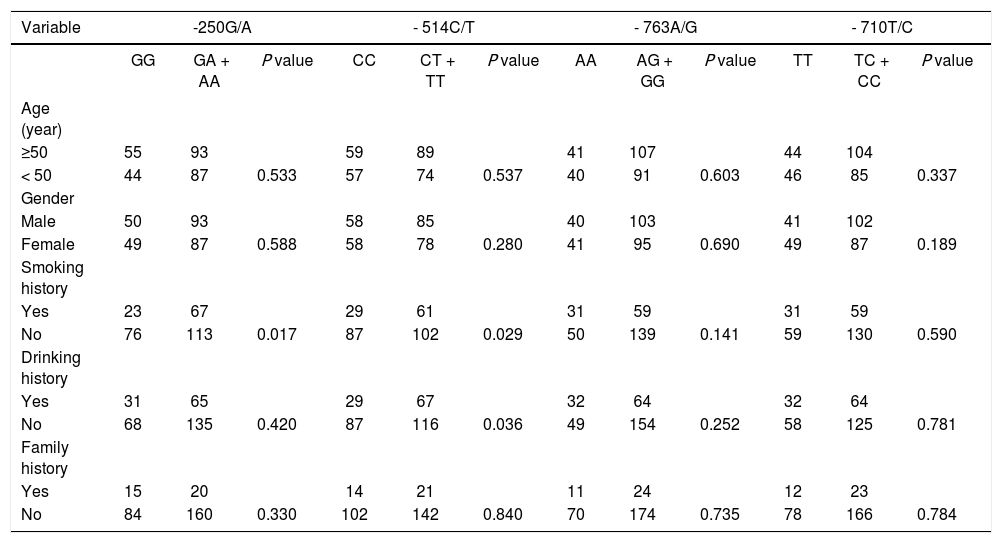
The associations between polymorphisms of -250G/A, -514C/T, -763A/G, -710T/C and pathological features indicated that the frequency of smoking in patients with GA + AA genotype in -250G/A was significantly higher than patients with GG genotype (p < 0.05), and there was no association between different genotypes in -250G/A and age, gender, drinking history, and family history of patients (all p > 0.05). The frequencies of smoking and drinking in patients with CT + TT genotype in -514C/T was significantly higher than those of with CC genotype (p < 0.05), and there was no association between different genotypes in -514C/T and the age, gender and family history of patients (all p > 0.05). And there was no association between different genotypes in -763A/G or -710T/C with the age, gender, smoking history, drinking history, and family history of patients (all p > 0.05) (Table 5). Results indicate that the GA + AA genotype in -250G/A and CT + TT genotype in -514C/T were associated with increased frequencies of smoking and drinking in HCC patients.
Associations between polymorphisms of four HL gene promoters and pathological features of HCC.
| Variable | -250G/A | - 514C/T | - 763A/G | - 710T/C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA + AA | Ρ value | CC | CT + TT | Ρ value | AA | AG + GG | Ρ value | TT | TC + CC | Ρ value | |
| Age (year) | ||||||||||||
| ≥50 | 55 | 93 | 59 | 89 | 41 | 107 | 44 | 104 | ||||
| < 50 | 44 | 87 | 0.533 | 57 | 74 | 0.537 | 40 | 91 | 0.603 | 46 | 85 | 0.337 |
| Gender | ||||||||||||
| Male | 50 | 93 | 58 | 85 | 40 | 103 | 41 | 102 | ||||
| Female | 49 | 87 | 0.588 | 58 | 78 | 0.280 | 41 | 95 | 0.690 | 49 | 87 | 0.189 |
| Smoking history | ||||||||||||
| Yes | 23 | 67 | 29 | 61 | 31 | 59 | 31 | 59 | ||||
| No | 76 | 113 | 0.017 | 87 | 102 | 0.029 | 50 | 139 | 0.141 | 59 | 130 | 0.590 |
| Drinking history | ||||||||||||
| Yes | 31 | 65 | 29 | 67 | 32 | 64 | 32 | 64 | ||||
| No | 68 | 135 | 0.420 | 87 | 116 | 0.036 | 49 | 154 | 0.252 | 58 | 125 | 0.781 |
| Family history | ||||||||||||
| Yes | 15 | 20 | 14 | 21 | 11 | 24 | 12 | 23 | ||||
| No | 84 | 160 | 0.330 | 102 | 142 | 0.840 | 70 | 174 | 0.735 | 78 | 166 | 0.784 |
HL: hepatic lipase.
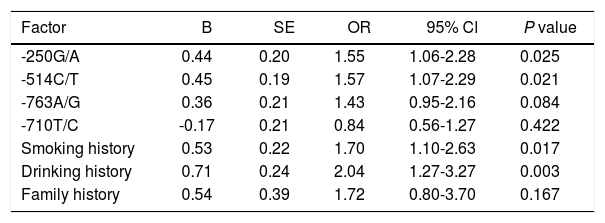
In order to investigate the risk factors of HCC, the logistic regression analysis was performed. The binary logistic regression analysis with the Enter model was performed with smoking history, drinking history, family history, and different genotypes of -250G/A, -514C/T, -763A/G, -710T/C as the independent variables, with the fact whether patients suffered from HCC as the dependent variable. It showed that -250G/A and -514C/T polymorphism, smoking history and drinking history of patients were significantly correlated with the risk of HCC, and -250G/A (GA + AA) (p = 0.025; OR = 1.55; 95%CI: 1.06-2.28), -514C/T (CT + TT) (p = 0.021; OR = 1.57; 95%CI: 1.07-2.29), smoking history (p = 0.017; OR = 1.70; 95%CI: 1.10-2.63) and drinking history (p = 0.003; OR = 2.04; 95%CI: 1.27-3.27) were demonstrated to be the risk factors for HCC (Table 6). These data indicate that polymorphisms at -250G/A and -514C/T, smoking history and drinking history were notably associated with the risk of HCC.
Multivariate logistic regression analysis for risk factors of HCC.
| Factor | B | SE | OR | 95% Cl | P value |
|---|---|---|---|---|---|
| -250G/A | 0.44 | 0.20 | 1.55 | 1.06-2.28 | 0.025 |
| -514C/T | 0.45 | 0.19 | 1.57 | 1.07-2.29 | 0.021 |
| -763A/G | 0.36 | 0.21 | 1.43 | 0.95-2.16 | 0.084 |
| -710T/C | -0.17 | 0.21 | 0.84 | 0.56-1.27 | 0.422 |
| Smoking history | 0.53 | 0.22 | 1.70 | 1.10-2.63 | 0.017 |
| Drinking history | 0.71 | 0.24 | 2.04 | 1.27-3.27 | 0.003 |
| Family history | 0.54 | 0.39 | 1.72 | 0.80-3.70 | 0.167 |
In Table 6, we have included -250G/A, -514C/T, -763A/G, -710T/C, smoking history, drinking history, and family history as independent variables to conduct binary logistic regression analysis with the Enter model to screen the factors affecting HCC risk; 250G/A: GG = 0, GA + AA = 1; -514C/T: CC = 0, CT + TT = 1; -763A/G: AA = 0; AG + GG = 1; -710T/C: TT = 0, TC + CC = 1; smoking history: without = 0, with = 1; drinking history: without = 0, with = 1; family history: without = 0, with = 1; the smoking history, drinking history, family history and other related environmental risk factors were analyzed to adjust the genotype regression results; SE: standard error. OR: odds ratio. CI: confidence interval. HCC: hepatocellular carcinoma.
Considered as one of the most prevalent and lethal malignancies worldwide, HCC is a common digestive system tumor with poor treatment effect and prognosis,15,16 which mainly prevails in Asia, especially in China.17 Previously, it has been reported that there are differences existed in genetic susceptibility to cancer among individuals, which are mediated by both environmental and genetic factors.18 Furthermore, various gene polymorphisms have been identified to be correlated with the occurrence and development of HCC.1,19 Therefore, this study was performed to investigate the association between the HL gene polymorphisms and risks of HCC in Chinese populations. Our results demonstrated that the -250G/A and -514C/T polymorphisms in HL gene promoter are associated with the risk of HCC.
The results of this study showed that compared to patients with GG wildtype of -250G/A, the risk of HCC in patients with GA and AA mutants increased prominently, and the risk was also higher in patients with CT and TT mutants in -514C/T than those with CC wildtype, which indicated that -250G/A and -514C/T polymorphisms in HL gene promoters are correlated with the risk of HCC. It has been reported that individuals were susceptible to developing certain cancers owing to genetic polymorphisms or alteration.18 For example, the polymorphic locus rs1047781 of FUT2 gene is correlated with HCC development, and FUT2 polymorphisms are proposed as predictors for tumor aggression in HCC.20 Based on the study of Leonhardt, et al., the PCR products of HL were identified in all the included HCC patients and healthy controls.21 As a liver-secreted lipolytic enzyme, HL is associated with the metabolism of plasma lipids that exist in high-density lipoproteins (HDL), low density lipoprotein (LDL) and in the very low density lipoprotein (VLDL).22,23 -250G/A, -514C/T, -763A/G, and -710T/C that have been identified as the four HL gene promoter polymorphisms are in complete linkage disequilibrium, which were reported to decrease the LIPC activity.24 Faggin, et al. have demonstrated that the common -514C/T variant in the HL gene promoter is associated with different levels of HL activity, LDL buoyancy, and HDL-2 cholesterol, in particular, compared with the CT and TT genotypes, the CC genotype is associated with higher HL activity, more atherogenic LDL particles and lower HDL cholesterol levels.25 Consistently, Jiang et al. have reported that the LDL-C levels in HCC patients were significantly lower than those in health people,26 and it was further reported that HL activity reduced remarkably when treated with intensive lipid-lowering therapy, which indicates that the HL activity and LDL level are positively related.27 In addition, it was evidenced that HL activity was higher in subjects with the GG genotype of HL gene promoter -250G/A than in those with the GA or AA genotypes.28
Besides, it was demonstrated in our study that HL gene promoter -250G/A and -514C/T polymorphisms and the habits of smoking and drinking were closely related to the risk of HCC. A previous study reported that alcohol intake and cigarette smoking are regarded as the two co-factors in the pathogenesis of HCC.29 Compared with nonsmokers, cigarette smokers are more likely to develop HCC.30 Alcohol intake is also reported to be tightly related to HCC, and there is a high mortality rate of smoker HCC patients.6
In conclusion, we found that the -250G/A and -514C/T polymorphisms in HL gene promoter are associated with the risk of HCC, while the polymorphisms o f -763A/G and -710T/C exerted no effect on the risk of HCC. In the view of the small sample size at the primary stage of our study, results may suffer from some bias. We also realize that the increased risk of HCC requires for the simultaneous changes and synergistic effects of multiple genes. Therefore, follow-up study will be needed to verify the association between -250G/A and -514C/T polymorphisms of HL gene and the risk of HCC, in order to lay a theoretical foundation for better prevention, diagnosis, and treatment measures for HCC at the genetic level.
Abbreviations- •
CI: confidence intervals.
- •
EDTA: elhylene diamine tetraacetic acid.
- •
HCC: hepatocellular carcinoma.
- •
HDL: high-density lipoproteins.
- •
HL: hepatic lipase.
- •
HSPG: heparan sulfate proteoglycans.
- •
HWE: Hardy-Weinberg equilibrium.
- •
PCR-RFLP: polymerase chain reaction restriction fragment length polymorphisms.
- •
SNPs: single nucleotide polymorphisms.
We would like to acknowledge the helpful comments on this paper received from our reviewers.
Conflict of InterestThe authors declares that there is no conflict of interest regarding the publication of this article.