Introduction and aim. Hepatitis B virus (HBV) infection remains a public health problem worldwide. In addition, HBV infection results are influenced by various virological, immunological, and genetic factors. Inducible T-cell costimulator (ICOS) polymorphisms involving chronic HBV infection have been confirmed in previous studies. This study was to explore the effects of ICOS single nucleotide polymorphisms in HBV subtypes and their interactions with viral mutations on HBV infection outcomes.
Material and methods. A total of 1,636 Han Chinese individuals were recruited, including 47 asymptomatic HBV carriers (ASC), 353 chronic hepatitis B (CHB) patients, 327 HBV-related liver cirrhosis (LC) patients, 193 HBV-related hepatocellular carcinoma (HCC) patients, 464 patients with spontaneous recovery from HBV infection (SR), and 252 healthy controls (HC). DNA samples from these subjects were genotyped for four ICOS SNPs (rs11883722, rs10932029, rs1559931, and rs4675379). Direct sequencing was used to determine the HBV mutations in the enhancer II, basal core promoter, and pre-core regions.
Results. We found that the genotype “TC” of ICOS rs10932029 SNP was associated with decreased HBV-related LC risk in the genotype C group. Additionally, the A1762T, G1764A and A1762T/G1764A mutations were associated with an increased risk of LC in the genotype C group. Further study indicated that interactions between ICOS rs10932029 genotype “TC” and A1762T or A1762T/G1764A mutations significantly decreased the LC risk in the genotype C group.
Conclusion. The rs10932029 genotype “TC” might be an LC-protective factor for HBV genotype C infection. The interactions between the rs10932029 genotype “TC” and A1762T or A1762T/G1764A mutations could decrease the risk of LC.
Hepatitis B virus (HBV) infection is prevalent around the world, especially in China. Almost 3 billion people are exposed to HBV, of which 350-400 million people are persistently infected worldwide. Approximately 93 million individuals are chronic HBV carriers, and the number of deaths from HBV-related liver disease has reached almost 300,000 per year in China.1 The final consequences of HBV infection vary widely in diverse individuals.For example, one HBV-infected individual may have normal liver function as an ASC, which has no effect on quality of life. While a patient with LC may develop many potentially lethal complications, such as ascites, gastrointestinal haemorrhage and hepatic encephalopathy. Therefore, we divide the patients into different sub-groups, including spontaneous recovery from HBV infection (SR), transition to an asymptomatic HBV carrier (ASC), chronic HBV (CHB), HBV-related liver cirrhosis (LC), HBV-related hepatocellular carcinoma (HCC). The mechanisms of HBV infection are complex procedures which are related to various virological, immunological, and host genetic factors.2 The initiation of the immune response is closely related to costimulatory molecule activation, which includes CD28 and the B7 family. ICOS is a member of the CD28 family, which is expressed on activated T-lymphocytes and forms homodimers to regulate cell-cell signalling, immune responses, and cell proliferation, and it has very important functions in T-lymphocyte-dependent humoural immunity.3–7 ICOS single nucleotide polymorphisms (SNPs) may affect protein expression and function. The function of ICOS in immune responses was described in our previous study, in which we determined that four ICOS SNPs, including rs11883722 in the promoter region, rs10932029 in intron 1, and rs1559931 and rs4675379 in the 3’-untranslated region (3’-UTR), are associated with viral clearance, persistent carriers after HBV infection, and LC/HCC development.8 However, the influence of combined ICOS SNPs with HBV genotypes and viral mutations is unclear. A previous study found several mutations in the EnhII/BCP/PC region of HBV genotypes B and C that were associated with HBV-related LC and HCC.9 The relationships between ICOS SNPs with mutations in the EnhII/BCP/PC and LC and HCC are undefined. Therefore, the aim of this study was to explore the effects of ICOS polymorphisms on HBV infection outcomes in HBV subtypes and their interactions with HBV mutations on HBV infection outcomes in a Han Chinese population.
Material and MethodsSubjectsA total of 1,636 patients were recruited from the Second Affiliated Hospital of Chongqing Medical University, Chongqing, China, from January 2011 through July 2014. All participants were hereditarily unrelated in a Han Chinese population. These subjects were divided into three groups: 464 patients with natural HBV clearance, 252 healthy controls (HC), and 920 chronic HBV patients (CH)(47 asymptomatic HBV carriers (ASC), 353 chronic hepatitis B patients (CHB), 327 HBV-related liver cirrhosis patients (LC), and 193 patients with HBV-related hepatocellular carcinoma (HCC) included). The demographics and laboratory characteristics of these subjects are listed in table 1. This research was approved by the Chongqing Medical University Second Affiliated Hospital Ethics Committee, and all participants provided written informed consent.
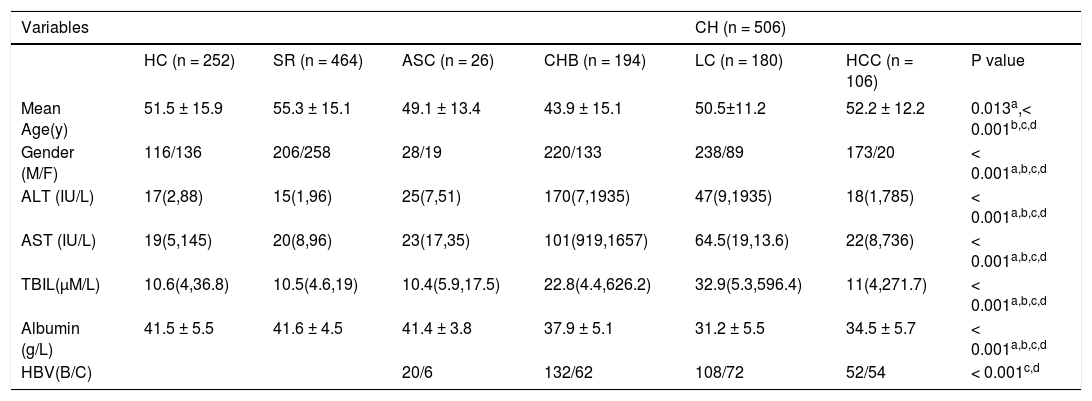
Characteristics of all subjects in this study
| Variables | CH (n = 506) | ||||||
|---|---|---|---|---|---|---|---|
| HC (n = 252) | SR (n = 464) | ASC (n = 26) | CHB (n = 194) | LC (n = 180) | HCC (n = 106) | P value | |
| Mean Age(y) | 51.5 ± 15.9 | 55.3 ± 15.1 | 49.1 ± 13.4 | 43.9 ± 15.1 | 50.5±11.2 | 52.2 ± 12.2 | 0.013a,< 0.001b,c,d |
| Gender (M/F) | 116/136 | 206/258 | 28/19 | 220/133 | 238/89 | 173/20 | < 0.001a,b,c,d |
| ALT (IU/L) | 17(2,88) | 15(1,96) | 25(7,51) | 170(7,1935) | 47(9,1935) | 18(1,785) | < 0.001a,b,c,d |
| AST (IU/L) | 19(5,145) | 20(8,96) | 23(17,35) | 101(919,1657) | 64.5(19,13.6) | 22(8,736) | < 0.001a,b,c,d |
| TBIL(µM/L) | 10.6(4,36.8) | 10.5(4.6,19) | 10.4(5.9,17.5) | 22.8(4.4,626.2) | 32.9(5.3,596.4) | 11(4,271.7) | < 0.001a,b,c,d |
| Albumin (g/L) | 41.5 ± 5.5 | 41.6 ± 4.5 | 41.4 ± 3.8 | 37.9 ± 5.1 | 31.2 ± 5.5 | 34.5 ± 5.7 | < 0.001a,b,c,d |
| HBV(B/C) | 20/6 | 132/62 | 108/72 | 52/54 | < 0.001c,d |
ALT: alanine aminotransferase. AST: aspartate aminotransferase. TBIL: serum total bilirubin. ASC: asymptomatic carriers. CHB: chronic hepatitis B. LC: liver cirrhosis. HCC: hepatocellular carcinoma. Values are shown as mean ± SD or median (range). Data were analyzed by binary logistic regression and adjusted for age and sex. a: between CH and HC. b: between CH and SR. c: between ASC + CHB and LC + HCC. d: between ASC + CHB + LC and HCC.
Selection criteria:
- •
The included subjects with HBV clearance were negative for HBsAg and HBV-DNA but positive for anti-HBs and anti-HBc and had normal alanine aminotransferase (ALT) and aspartate transaminase (AST) levels,which are analyzed by automatic biochemical analyzer(Hitach,Japan),the normal rang usually considered to be 5 to 40 U/L.
- •
The HC subjects were HBsAg, HBeAg, anti-HBs, anti-HBe and anti-HBc negative and were never exposed to HBV.
- •
The ASC patients were seropositive for HBsAg without any clinical liver disease and with normal ALT and/ or AST levels.
- •
The CHB subjects were diagnosed based on their HBsAg seropositivity, positivity for serum HBV-DNA levels, and continuously elevated ALT levels over a period of 6 months, and they did not receive antiviral therapy before we collected their blood samples. The ALT and/or AST levels in the CHB group were two times higher than the upper limits of the normal range, but they were free of imageological examination or histologic cirrhosis and HCC at diagnosis.
- •
The LC group was diagnosed by gastroscopy and abdominal ultrasound examinations and/or computed tomography (CT), and the participants who demonstrated explicit signs of cirrhosis and who were free of HCC were diagnosed with LC.
- •
HCC was diagnosed by elevated alpha-fetoprotein (AFP) and imageological examination or liver tissue histology.
Participants were excluded from this research if they met any of the following exclusion criteria:
- •
Any other hepatic viral infection (HAV, HCV, HDV or HEV) or human immunodeficiency virus infection.
- •
Other chronic liver diseases, such as autoimmune liver disease, drug-induced hepatic diseases or alcoholic liver diseases.
- •
Serious cardiovascular, respiratory or renal system diseases.
- •
Autoimmune diseases or tumors in other organs.
- •
18 years of age or under, pregnancy or not of Han origin.
The methods and processes of choosing these four different ICOS SNPs (rs11883722, rs10932029, rs1559931, and rs4675379) were described in our previous study.8
SNP DNA extraction and genotypingFive milliliter of fasting venous blood was collected in ethylenediaminetetraacetic acid containing tubes from each participant. Genomic DNA was segregated from the leukocytes of 200 µL of venous blood using a Wizard® Genomic DNA Purification Kit according to the manufacturer’s instruction (Promega, USA). The DNA concentration was measured by a NanoDrop spectrophotometer and diluted to 20 ng/µL and then stored at -80°C for future analysis. We used a predesigned TaqMan assay and an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) to genotype the four SNPs.8
HBV extraction, genotyping and mutation analysisWe utilized viral genome extraction kits (Biomed, Beijing, China) to extract the HBV DNA from the chronic HBV patients. The fragment between positions 1,604 to 2,006 (including the EnhII region [nucleotides 1,6851,773], the BCP region [nucleotides 1,742-1,849], and the pre-core region [nucleotides 1,814-1,901]) of the HBV were amplified by nested PCR. The first-round PCR of positions 1,604-2,006 was performed in a 20-µL reaction mixture with primers 1,604F (5’-TCGCATGGAGACCACCGTGA-3’) and 2072R (5’-CCTGAGTGCTGTATG GTGAG-3’) with a 3-min hot start followed by 35 cycles of 95°C for 25 s, 57°C for 25 s, and 72°C for 60 s. The second-round PCR was performed on 0.2 µL of the first-round product in a 20-µL reaction mixture with primers 1604F (5’-TCGCATGGAGACCACCGTGA-3’) and 2,006R (5’-ATACAGAGCAGAGGCG GTAT-3’) with a 3-min hot start, followed by 30 cycles of 95°C for 25 s, 57°C for 25 s, and 72°C for 50 s. An ABI 3,730 Genetic Analyser (Applied Biosystems, Foster City, CA) was used for direct DNA sequencing. To confirm the accuracy of the geno-typing results, we used real-time PCR to determine the HBV genotypes and an NCBI genotyping tool (www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi) for verification.
Wild-type and mutated nucleotide definitionsBased on the standard of greater than 8% diversity in the whole HBV genome, HBV has at least 8 genotypes,10–12 and the newer genotypes I and J have been identified in recent years.13,14 Nevertheless, HBV genotypes have distinct geographic distributions, and genotypes B/C are endemic in East Asia.11 Wild-type and mutated nucleotides of HBV were defined in genotypes B and C groups. Because the wild-type strain traditionally comes from HBeAg-positive patients,15,16 we defined the highest frequency among ASC patients with seropositivity for HBeAg as a wild-type nucleotide. HBV mutations were defined as a substitution at each site with other nucleotides. Mutations of the “hot spot” type were defined as sites with a frequency >10% in all HBV-infected subjects.
Statistical analysisThe Hardy-Weinberg equilibrium (HWE) for each genotype was tested online. We analysed the SNPs’ associations with HBV susceptibility, clearance and chronic infection in the HBV genotype B/C groups. By comparing the different groups, which is helpful to understand the essence of ICOS SNPs in HBV persistence, HBV clearance, and diseases,have yet to be confirmed in our previous study.8 By contrast, in the HCs and the infected cases (SR + CH), we explored the associations between SNPs and HBV susceptibility. Additionally, we investigated the connection between the SR and the CH patients with the removal of infection. In the HBV infection subtypes, we did not use contrastive analysis between the CHB and ASC patients because of the lack of ACS cases. At the same time, the CHB cases were not life-threatening, therefore, we regarded the ASC and CHB cases as control cases, in which we analysed the effect of SNPs and mutations in patients with HBV-related LC and HBV-related HCC. For most patients with HBV-related HCC have underlying cirrhosis. Therefore, we separated the HBV-infected patients into four groups before we analysed the relationships between LC and HCC: LC (LC plus HCC), LC-free (ACS plus CHB), HCC and HCC-free (ASC, CHB plus LC). We adopted a binary logistic regression to analyse the influence of SNP polymorphisms on HBV susceptibility/clearance/progression in genotypes B or C groups after adjusting for age and gender. Disease characteristics (such as gender and AST and albumin levels) are expressed as the mean ± standard deviation or median (range) and we compared them using Student’s t test, χ2 test or Mann-Whitney U test. All tests were two-sided, and a p value ≤ 0.05 was considered statistically significant. The data were analysed with the SPSS 19.0 data analysis software package (SPSS, Chicago, IL, USA).
ResultsCharacteristics of the study populationAs shown in table 1, the gender proportion differed significantly amongst the four chronic HBV patient groups. The percentage of men enrolled in this study was obviously higher in the infection group (including ASC, CHB, LC, and HCC) than in the HC and HBV clearance groups, a finding that was similar to a previous epidemiological survey.16 The ages of the patients in the four chronic HBV patient groups were also significantly different, which may represent differences in disease processes. The ALT, AST, TBIL and albumin levels in the six groups presented with different HBV infection processes and statuses. The serological analyses indicated that the subjects we recruited met the appropriate diagnostic standards.
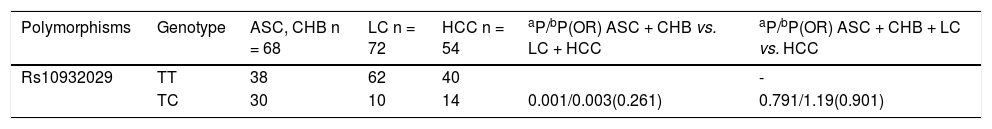
Correlations between ICOS SNP polymorphisms and HBV infection subtype outcomesWe found that ICOS rs1183722 did not conform to HWE; therefore, we did not further analyse that SNP. In our present study, we analysed the relevant research of ICOS SNPs with several HBV genotype B or C infection outcomes. There were no significant associations between the SNPs and spontaneous HBV clearance (SR vs. CH). The correlations between the ICOS SNPs and HBV susceptibility (HC vs. CH plus SR) were also not significantly different. The ICOS rs10932029 SNP (TC vs. TT) was associated with decreased HBV-related LC risk (LC plus HCC) only with HBV genotype C infection (P = 0.003, OR = 0.261) compared with the LC-free HBV-infected patients (ASC plus CHB) in table 2. However, no ICOS SNPs influenced the HBV-related LC for genotype B, and no SNP had a consistent effect on the HBV genotype B or C infection regarding HCC risk.
Multivariate analysis of polymorphisms of three ICOS SNPs (rs10932029, rs1559931, and rs4675379) in chronic HBV genotype C infection.
| Polymorphisms | Genotype | ASC, CHB n = 68 | LC n = 72 | HCC n = 54 | aP/bP(OR) ASC + CHB vs. LC + HCC | aP/bP(OR) ASC + CHB + LC vs. HCC |
|---|---|---|---|---|---|---|
| Rs10932029 | TT | 38 | 62 | 40 | - | |
| TC | 30 | 10 | 14 | 0.001/0.003(0.261) | 0.791/1.19(0.901) |
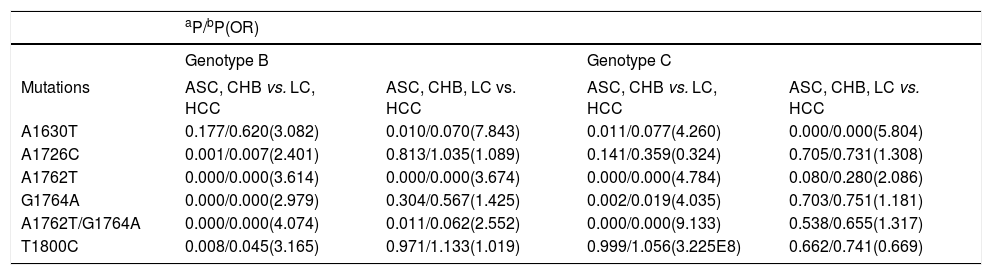
DNA samples were sequenced for 506 (55%) of the HBV-infected subjects in the HBV EnhII/BCP/PC region. The “hot spot” mutations in the EnhII/BCP/PC region were found at nt. 1630, 1632, 1633, 1635, 1636, 1637, 1638, 1652, 1653, 1753, 1679, 1726, 1727, 1730, 1752, 1753, 1757, 1762, 1764, 1766, 1799, 1800, 1846, 1896 and 1899. Additionally, the “hot spot” mutations included simultaneous nt. 1762 and 1764 mutations, which was similar to a previous discovery.17 As shown in Table 3, we investigated several LC- or HCC-related HBV mutations in B and C. We found that A1630G, A1726C, A1762T, G1764A, A1762T/ G1764A and T1800C were LC- or HCC-related mutations for genotype B or C. The A1726C (P = 0.007, OR = 2.401), A1762T (P = 0.000, OR = 3.614), G1764A (P = 0.000, OR = 2.979), A1762T/G1764A (P = 0.000, OR = 4.074) and T1800C mutations (P = 0.045, OR = 3.165) were LC-risk mutations for genotype B when the LC patients (LC plus HCC) were compared with the LC-free patients (ASC plus CHB). In addition, the A1762T mutation (P = 0.000, OR = 3.674) was an HCC-risk mutation for genotype B when HCC patients were compared with HCC-free patients (ASC plus CHB and LC). The n1762 mutations were not obviously different between the LC and HCC patients in HBV genotype B infections. The A1762T (P = 0.000, OR = 4.784), G1764A (P = 0.019, OR = 4.035) and A1762T/G1764A mutations (P = 0.000, OR = 9.133) were associated with an increased LC risk in HBV genotype C infections. The A1630G mutation (P = 0.000, OR = 5.804) was associated with HCC risk for genotype C.
Associations of mutations with LC and HCC in genotype B or C.
| aP/bP(OR) | ||||
|---|---|---|---|---|
| Genotype B | Genotype C | |||
| Mutations | ASC, CHB vs. LC, HCC | ASC, CHB, LC vs. HCC | ASC, CHB vs. LC, HCC | ASC, CHB, LC vs. HCC |
| A1630T | 0.177/0.620(3.082) | 0.010/0.070(7.843) | 0.011/0.077(4.260) | 0.000/0.000(5.804) |
| A1726C | 0.001/0.007(2.401) | 0.813/1.035(1.089) | 0.141/0.359(0.324) | 0.705/0.731(1.308) |
| A1762T | 0.000/0.000(3.614) | 0.000/0.000(3.674) | 0.000/0.000(4.784) | 0.080/0.280(2.086) |
| G1764A | 0.000/0.000(2.979) | 0.304/0.567(1.425) | 0.002/0.019(4.035) | 0.703/0.751(1.181) |
| A1762T/G1764A | 0.000/0.000(4.074) | 0.011/0.062(2.552) | 0.000/0.000(9.133) | 0.538/0.655(1.317) |
| T1800C | 0.008/0.045(3.165) | 0.971/1.133(1.019) | 0.999/1.056(3.225E8) | 0.662/0.741(0.669) |
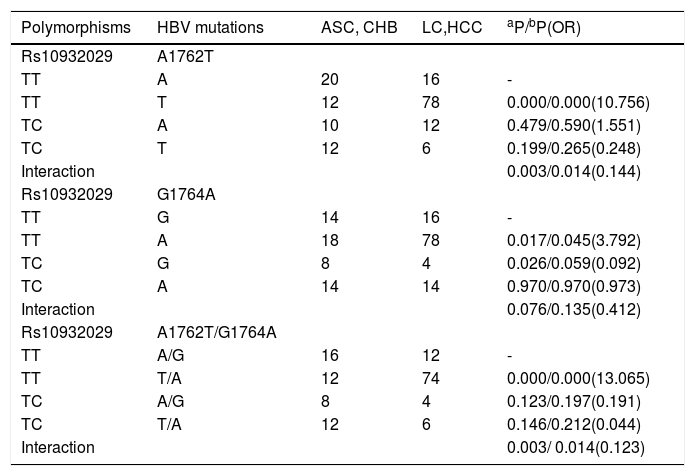
We surveyed multiplicative interactions between rs10932029 and all LC-related mutations (T1762T, G1764A and A1762T/G1764A) for genotype C. Interestingly, the rs10932029 genotype “TC” was an LC-protective factor; however, the LC-related mutations were LC-risk factors for genotype C groups. Table 4 presented the effects of interactions between rs10932029 (TC vs. TT) and LC-risk mutations in the EnhII/BCP/PC region for genotype C. Interactions between ICOS rs10932029 (TC vs.TT) and A1762T (P = 0.014, OR = 0.144) or A1762T/G1764A (P = 0.014, OR = 0.123) mutations significantly decreased the LC risk for genotype C when the LC patients (LC plus HCC) were compared with the LC-free patients (ASC plus CHB). However, the interaction between ICOS rs10932029 (TC vs. TT) and G1764A mutation (P = 0.076) was not significantly associated with LC. This finding verified that the HBV infection outcomes were concurrently influenced by various virological and host genetic factors. Moreover, when various virological and host genetic factors exerted opposite effects on the chronic HBV infection progression, the final consequences are disparate.
Interactions of ICOS rs10932029 with the LC-related HBV mutations in genotype C HBV-infected individuals.
| Polymorphisms | HBV mutations | ASC, CHB | LC,HCC | aP/bP(OR) |
|---|---|---|---|---|
| Rs10932029 | A1762T | |||
| TT | A | 20 | 16 | - |
| TT | T | 12 | 78 | 0.000/0.000(10.756) |
| TC | A | 10 | 12 | 0.479/0.590(1.551) |
| TC | T | 12 | 6 | 0.199/0.265(0.248) |
| Interaction | 0.003/0.014(0.144) | |||
| Rs10932029 | G1764A | |||
| TT | G | 14 | 16 | - |
| TT | A | 18 | 78 | 0.017/0.045(3.792) |
| TC | G | 8 | 4 | 0.026/0.059(0.092) |
| TC | A | 14 | 14 | 0.970/0.970(0.973) |
| Interaction | 0.076/0.135(0.412) | |||
| Rs10932029 | A1762T/G1764A | |||
| TT | A/G | 16 | 12 | - |
| TT | T/A | 12 | 74 | 0.000/0.000(13.065) |
| TC | A/G | 8 | 4 | 0.123/0.197(0.191) |
| TC | T/A | 12 | 6 | 0.146/0.212(0.044) |
| Interaction | 0.003/ 0.014(0.123) |
ICOS SNPs are associated with HBV infection, HBV clearance, and the progression of persistent HBV infection.8 However, prior studies did not consider the effect of different HBV subtypes on these outcomes. To comprehensively analyse this finding, we investigated several associations between HBV genotype B and C infections. This was the first description of an association between HBV subtypes and their interactions with viral mutations of HBV infection outcomes in a Han Chinese population. As such, we elucidated that the genotype “TC” of rs10932029 plays a protective role in HBV-related LC risk in HBV genotype C infection. A previous study also determined that an NFKB polymorphism is related to gene expression regulation.18 An earlier research study found that rs10932029 is associated with gene expression regulation (such as mRNA half-life and translation efficiency);19 therefore, we speculate that rs10932029 may influence the HBV-related LC process. The rs10932029 of ICOS has also been associated with breast cancer risk.20 A previous study showed that the ICOS rs1559931 SNP is associated with decreased HBV-related HCC risk,8 although we did not observe this relationship. This discrepancy was apparent because of the different HBV subtype frequency. A Japanese group observed that the percentages of different HBV genotype infections in HCC patients were different (48.9% of HCC patients had HBV C, 6.7% had HBV Ba, and 40.0% had HBV Bj) in the Tohoku district and in Okinawa.21 Additionally, complex genes or environmental factors may also be possible causes. Therefore, further studies are needed to make after subdividing the HBV subgenotype B infections.
To investigate the interactions between the ICOS SNPs and viral mutations with HBV infection outcomes, we analysed the associations between viral mutations and LC and HCC in the HBV B and C EnhII/BCP/PC region. The results indicated that the functions of the same mutations in the HBV infection progress may be different in different HBV genotypes. A1726C and T1800C mutations increased the LC risk only in genotype B groups. The A1630G mutation is an HCC-risk mutation only for HBV C infection. The A1762T and G1764A mutations and A1762T/G1764A double mutations increased the LC-risk for both genotypes B and C infection. A previous study indicated that A1762T and G1764A were significantly associated with LC for HBV genotype B and C infections.22 The A1726T mutation simultaneously increased the risk of LC and HCC;although, the mutation frequency was not significantly different between the LC and HCC cases.Wherefore, we elucidated that the A1726T mutation might result in unfavorable outcomes in chronic HBV-infected individuals. The increased HCC risk association with A1762T/G1764A double mutations have been verified in many studies,23,24 however, have no correlation to HCC in the current study. One reason for this discovery with has fewer HCC patients than HCC-free patients, which generated a result bias. Another reason is that some HCC-protective factors probably work at the same time, as mentioned above,did not take into account.
Another meaningful discovery is that the interactions between the rs10932029 genotype “TC” with the A1762T or A1762T/G1764A mutations decreased the LC risk. Nevertheless, the single rs10932029 genotype “TC” and the A1762T or A1762T/G1764A mutations had an inverse effect on the LC risk. The current study found that the single rs10932029 genotype “TC” and G1764A mutations had an effect on HBV-related LC alone, however, the interaction was not significantly different. These results verified that the HBV infection outcomes were affected by host genetic factors and viral mutations, and when these were opposite, the interactions were multiple. Considering that HBV mutations may be influenced by host immune functions to a certain extent,25 we speculate that host genetic factors might have a stronger influence than that of viral mutations (Table 4). Consequently, we observed no positive finding between ICOS SNPs and viral mutations.
This study reveals that the rs10932029 genotype “TC” might be an LC-protective factor in HBV genotype C infections. Additionally, the interactions between the rs10932029 genotype “TC” with A1762T or A1762T/ G1764A mutations could decrease the LC risk. ICOS is a member of T-cell molecule regulation family and localize at the chromosome region 2q33-3.26 ICOS proteins can form homodimers which can activate on T-lymphocytes and regulate cell proliferation and enhance cytokine secretion and humoral immunity in turn.3,27 ICOS activities may help the body to eradicate HBV infection. Therefore, a new way of thinking may be proposed for reducing the incidence of LC. This is the first study to clarify the interactions between ICOS SNPs with viral mutations and the influence of ICOS SNPs on HBV genotypes B and C infections. In addition, this study also has some limitations. First, the sample sizes were relatively small, which may led to rs11883722 genotype frequencies not conform to Hardy-Weinberg distribution. Therefore, we did not analyse rs11883722, which has been found to be associated with allergies, Th2 cytokine production, and serum IgE levels in members of the Hutterite lineage.28 Relatively small sample sizes might be one of the reasons why the LC- or HCC-related mutations in our study were fewer than others. Second, we did not subdivide the HBV genotype B into subgenotypes, because we observed a remarkable difference in the percentage of HCC patients with the HBV genotype B.21 Third, the host genes were too complicated to analyse synthetically, as other genes, including pre-miR-218 polymorphisms, CD8+ T cell inhibitory genes, STAT4 and HLA-DQ are affected during HBV infection.29–31 In view of this study was a cross-sectional study, we were able to learn about the connection between disease and specific population at some point. Because chronic HBV progression may develop into LC or HCC after several years, further research using a larger sample sizes and follow-up visits are needed, so that the disease progression of chronic HBV-infected patients can be evaluated long term.
Abbreviations- •
ALT: alanine aminotransferase.
- •
ASC: asymptomatic HBV carriers.
- •
AST: aspartate aminotransferase.
- •
CHB: chronic hepatitis B.
- •
HBV: hepatitis B virus.
- •
HC: healthy controls.
- •
HCC: hepatocellular carcinoma.
- •
ICOS: inducible T-cell costimulator.
- •
LC: liver cirrhosis.
- •
SNPs: Single nucleotide polymorphisms.
- •
SR: spontaneous recovery from HBV infection.
- •
TBIL: serum total bilirubin.
The authors declares that there is no conflict of interest regarding the publication of this article.