Hereditary sudden cardiac death syndromes comprise a wide range of diseases resulting from alteration in cardiac ion channels. Genes involved in these syndromes represent diverse mutations that cause the altered encoding of the diverse proteins constituting these channels, thus affecting directly the currents of the corresponding ions. In the present article we will briefly review how to arrive to a clinical diagnosis and we will present the results of molecular genetic studies made in Mexican subjects attending the SCD Syndromes Clinic of the National Institute of Cardiology of Mexico City.
Los síndromes hereditarios de muerte súbita cardíaca comprenden una amplia gama de enfermedades resultantes de la alteración en los canales iónicos cardíacos. Los genes implicados en estos síndromes presentan mutaciones que causan alteraciones de las diversas proteínas que constituyen estos canales y que, por lo tanto, afectan directamente a las diferentes corrientes iónicas. En el presente artículo se revisa brevemente la forma de llegar a un diagnóstico clínico de dichos síndromes y se presentan los resultados de los estudios genéticos moleculares realizados en sujetos mexicanos que asisten a la Clínica de Síndromes Hereditarios de Muerte Súbita del Instituto Nacional de Cardiología Ignacio Chávez.
Hereditary sudden cardiac death (SCD) syndromes comprise a wide range of diseases resulting from alteration in cardiac ion channels.1 Genes involved in these syndromes represent diverse mutations that cause an altered encoding of the diverse proteins constituting these channels, thus affecting directly the currents of the corresponding ions. An increase or a diminution in the flow of these ions through the cell membrane, markedly affects ventricular repolarization giving rise to severe (potentially fatal) ventricular arrhythmias (ventricular tachycardia, ventricular fibrillation or both).2–6 In the present article we will briefly review how to arrive to a clinical diagnosis and present the results of molecular genetic studies made in Mexican subjects attending the SCD Syndromes Clinic of the National Institute of Cardiology “Ignacio Chavez” Mexico City.
Clinical diagnosis of SCD syndromesClinical diagnosis of hereditary SCD syndromes is established when there is a family history of SCD or a personal history of aborted SCD. The 12-lead ECG usually has some of the characteristic signs, specific to each of the different channel diseases including a long (Long QT syndrome) or a short QT interval (Short QT syndrome), an image of right bundle branch block with ST segment elevation (Brugada syndrome), epsilon waves (Arrhythmogenic Right Ventricular Cardiomyopathy) or large U waves (Andersen–Tawil Syndrome), etc.7 The clinical diagnosis should be confirmed whenever possible by a molecular study. All family members should be screened to provide appropriate genetic counseling and help other possible affected members with the disease.
Even with the advance in genetic screening for cardiac ion channel diseases, the diagnosis is still clinical. Genealogy (family history) must be done by someone with enough expertise in order to obtain relevant information to SCD Syndromes. It is important not only to ask the subject, but also their relatives for obtaining a complete family history. This is especially important in case of youngsters or people with low-educational background. A detailed history using specific terms and their correspondent meanings must be explained to the patient. Specific questions could be: has someone in your family suddenly died? Has someone in your family died before the age of 45? Has someone in your family died because of a supposed heart attack? Has someone in your family been diagnosed as epileptic? Has someone in your family fainted?
In order to achieve a better clinical diagnosis when asking about the family history, the question cannot rely only on the answers provided by the patient or their relatives. It is very important to ask for official documents that could support the diagnosis. These documents usually exist in the hospitals where the final attention to the deceased relatives occurred, but many times nobody takes the trouble to collect and bring them to the physician to review the information. Our duty is to ask for them as they may help to elaborate the clinical diagnosis.
When asking the individual patient, one must be aware of how the presentation of the different syndromes occurred. Ask not only about the presence of syncope and pre-syncope, but also about the specific circumstances in which they occurred: the time of the day, activity performed at that moment, and the presence or absence of premonitory signs or symptoms. Sometimes the relatives or the person that witnessed the episode may provide this information. The physician should ask for true unconsciousness and how long it lasted. The possibility of epileptic activity (asking for proper signs and symptoms) should be explored. We must ask for agonic respiration. If there were witnesses, ask how they found the patient. If medical or paramedical personnel attended the patient, they might have reported how they found him or her. Looking for their first ECG tracings if available could be relevant as well as asking about all the procedures applied by those personnel.
The specific characteristics of each syndrome have been extensively reviewed by many groups and are not part of this review.8,9
Molecular diagnosis of SCD syndromesTogether with the complete clinical history, molecular genetic testing is an additional tool for the diagnosis of these syndromes. It can help to establish or confirm a clinical or presumptive diagnosis, and study relatives which is now called “cascade screening”. It is also useful to stratify the risk associated with specific mutations and therefore, to take therapeutic decisions such as the use of beta-blockers, referral for sympathetic denervation or implantation of an implantable cardioverter-defibrillator (ICD).
Detection of pathogenic mutation in family members should prompt regular cardiac evaluations with estimation of the risk of sudden cardiac death in each consultation. The objective is to initiate early medical treatment to prevent SCD in relatives affected by the mutation. This includes lifestyle advice (generally restricted from competitive sports, strenuous physical exercise or both), drugs or ICD implantation. For example, in ARVD, Hodgkinson et al.10 reported a subtype with a unique ECG finding (poor progression of R wave), high incidence of LV dilatation, heart failure or both and early death, particularly in males. In this subtype of ARVD, molecular pre-symptomatic diagnosis has the greatest clinical utility as adult males with the TMEM43 (transmembrane protein 43) p.S358L gene mutation may benefit from early ICD therapy. Bastarrika et al.11 reported a recent case of a male with some such characteristics although without genetic studies. It will certainly be very useful to perform genetic screening in that type of families in order to identify these “high-risk” mutations.
For subjects having this condition who wish to have children, reproductive counseling (“genetic counseling”) about the risk of transmission of the mutation to their offspring may be provided.
It is important to establish that in many diseases associated with SCD, genetic tests are not indispensable in the diagnosis because they do not influence the disease management. In consequence, and to help physicians to incorporate the results of genetic testing into their practice, the European Journal of Human Genetics has released a series of publications called “Clinical Utility Gene Cards”. The one by Te Rijdt et al.9 on ARVD is highly recommended.
Next-generation sequencing for the molecular diagnosis in Mexican subjects with SCD syndromesAlthough hereditary SCD syndromes have clinical and electrocardiographic characteristics that generally allow their differentiation, a molecular diagnosis of the underlying mutation is necessary for providing adequate genetic counseling for the rest of the family. Through collaboration between the Instituto Nacional de Cardiología “Ignacio Chávez” and a private enterprise (Sistemas Genómicos, Valencia, Spain), molecular analyses were performed in eight Mexican subjects with clinical diagnosis of a hereditary SCD syndrome.
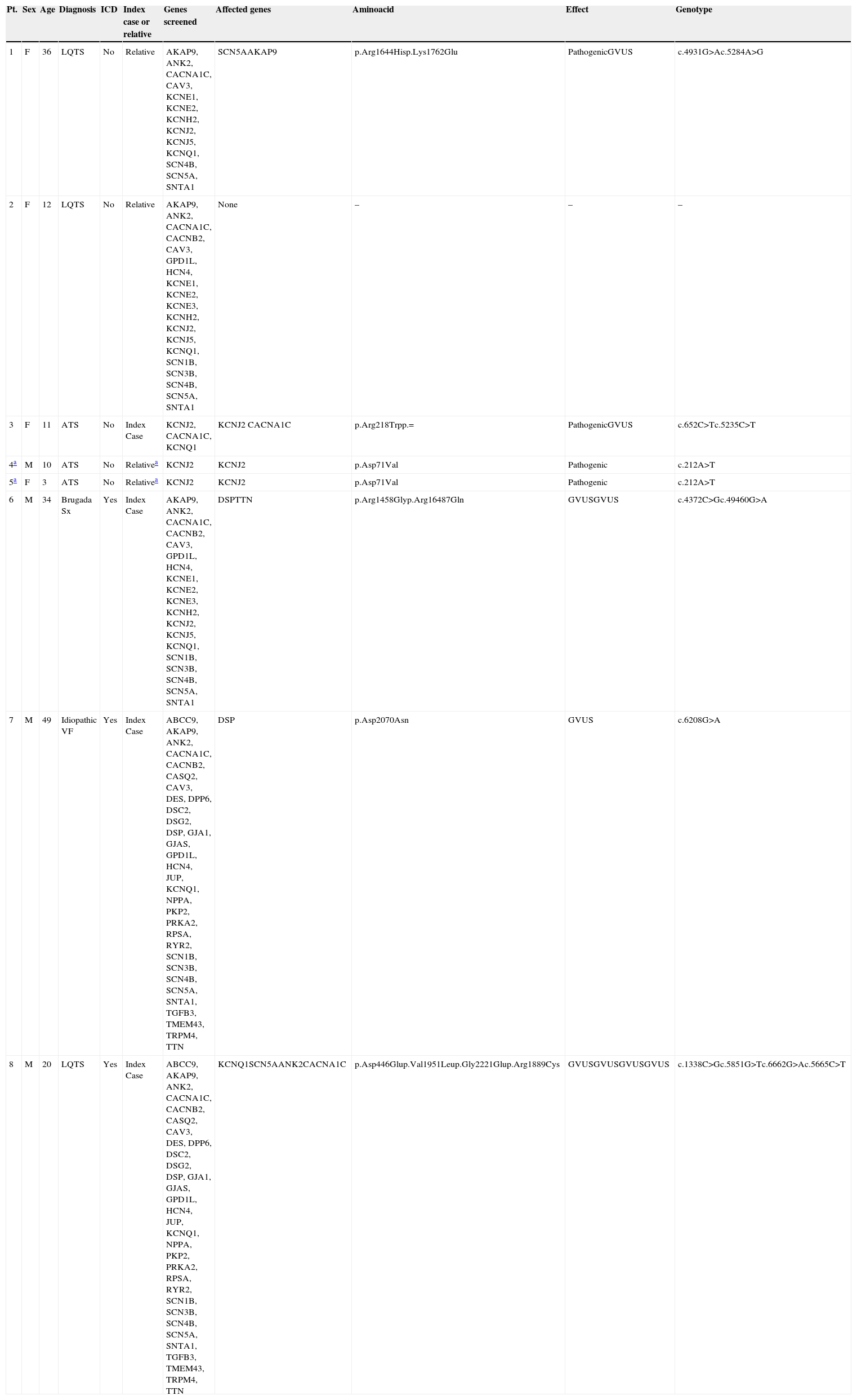
Table 1 shows the complete list of genes screened and the results of genetic testing of the subjects studied, who are either index cases (n=4) or relatives of index cases (n=4). The selection of genes to be analyzed by next generation sequencing (NGS) was based on the diagnosis or clinical suspicion.
Clinical characteristics of the subjects studied by next generation sequencing, including genes studied and affected. Exact mutations and its effect (pathogenic or not) are described.
| Pt. | Sex | Age | Diagnosis | ICD | Index case or relative | Genes screened | Affected genes | Aminoacid | Effect | Genotype |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 36 | LQTS | No | Relative | AKAP9, ANK2, CACNA1C, CAV3, KCNE1, KCNE2, KCNH2, KCNJ2, KCNJ5, KCNQ1, SCN4B, SCN5A, SNTA1 | SCN5AAKAP9 | p.Arg1644Hisp.Lys1762Glu | PathogenicGVUS | c.4931G>Ac.5284A>G |
| 2 | F | 12 | LQTS | No | Relative | AKAP9, ANK2, CACNA1C, CACNB2, CAV3, GPD1L, HCN4, KCNE1, KCNE2, KCNE3, KCNH2, KCNJ2, KCNJ5, KCNQ1, SCN1B, SCN3B, SCN4B, SCN5A, SNTA1 | None | – | – | – |
| 3 | F | 11 | ATS | No | Index Case | KCNJ2, CACNA1C, KCNQ1 | KCNJ2 CACNA1C | p.Arg218Trpp.= | PathogenicGVUS | c.652C>Tc.5235C>T |
| 4a | M | 10 | ATS | No | Relativea | KCNJ2 | KCNJ2 | p.Asp71Val | Pathogenic | c.212A>T |
| 5a | F | 3 | ATS | No | Relativea | KCNJ2 | KCNJ2 | p.Asp71Val | Pathogenic | c.212A>T |
| 6 | M | 34 | Brugada Sx | Yes | Index Case | AKAP9, ANK2, CACNA1C, CACNB2, CAV3, GPD1L, HCN4, KCNE1, KCNE2, KCNE3, KCNH2, KCNJ2, KCNJ5, KCNQ1, SCN1B, SCN3B, SCN4B, SCN5A, SNTA1 | DSPTTN | p.Arg1458Glyp.Arg16487Gln | GVUSGVUS | c.4372C>Gc.49460G>A |
| 7 | M | 49 | Idiopathic VF | Yes | Index Case | ABCC9, AKAP9, ANK2, CACNA1C, CACNB2, CASQ2, CAV3, DES, DPP6, DSC2, DSG2, DSP, GJA1, GJAS, GPD1L, HCN4, JUP, KCNQ1, NPPA, PKP2, PRKA2, RPSA, RYR2, SCN1B, SCN3B, SCN4B, SCN5A, SNTA1, TGFB3, TMEM43, TRPM4, TTN | DSP | p.Asp2070Asn | GVUS | c.6208G>A |
| 8 | M | 20 | LQTS | Yes | Index Case | ABCC9, AKAP9, ANK2, CACNA1C, CACNB2, CASQ2, CAV3, DES, DPP6, DSC2, DSG2, DSP, GJA1, GJAS, GPD1L, HCN4, JUP, KCNQ1, NPPA, PKP2, PRKA2, RPSA, RYR2, SCN1B, SCN3B, SCN4B, SCN5A, SNTA1, TGFB3, TMEM43, TRPM4, TTN | KCNQ1SCN5AANK2CACNA1C | p.Asp446Glup.Val1951Leup.Gly2221Glup.Arg1889Cys | GVUSGVUSGVUSGVUS | c.1338C>Gc.5851G>Tc.6662G>Ac.5665C>T |
ATS=Andersen–Tawil Syndrome, GVUS=genetic variant of unknown significance, ICD=implantable automatic defibrillator, VF=ventricular fibrillation, LQTS=long QT syndrome.
Technical details of NGS are as follows: Target regions were captured (SureSelect, Agilent), and sequenced in an Illumina MiSeq platform. Bioinformatic analysis consists of mapping and comparing reads against the reference sequence of the human genome, aligning them, identification of point variations and small indels, as well as their functional annotation. A series of filters were applied to the list of all annotated variants in order to catch the pathogenic mutation or obtain a few potentially pathogenic variants. The candidate variants were then thoroughly researched in the scientific literature and databases. For this, a broad set of sources was consulted (generic databases such as the HGMD Pro, ClinVar and LOVD; locus specific databases including the Sarcomere Protein Gene Mutation Database, ARVD/C mutation database, etc., as well as relevant bibliography via PubMed/MEDLINE). The Kaviar database, which groups together 57 variation resources (dbSNP135, HapMap, 1000Genomes, 200Exomes, etc.), was also used. In silico algorithms were used in case of novel missense changes (Align GVGD, SIFT, PolyPhen-2, Condel, MutationTaster, MAPP and Mutpred), or splice-site variants (NNSPLICE, Human Splicing Finder, MaxEntScan, GeneSplicer and SpliceSiteFinder-like). All this information were taken into account for ranking and classifying variants into: ‘pathogenic’, ‘probably pathogenic’, ‘variant of unknown significance (GVUS, genetic variant of unknown significance)’, ‘probably non-pathogenic’, or ‘non-pathogenic’ classes. Those variants classified as ‘pathogenic’, ‘possibly pathogenic’, or ‘variant of unknown significance’, undergo confirmation using Sanger sequencing.
A molecular result was attained in seven out of eight patients. Only in one girl with long QT syndrome it was not possible to obtain a result despite having analyzed 19 genes associated with SCD. We detected four mutations with pathogenic significance and nine GVUS. All mutations detected by NGS were confirmed through Sanger sequencing. A Long QT Syndrome was confirmed in a woman in whom a pathogenic mutation was documented in SCN5A gene, which codes for the sodium channel. In all Andersen–Tawil syndrome cases a mutation of the potassium KCNJ2 channel was found. This was previously reported by Plaster et al.12. We also found mutations classified as GVUS accompanying two cases of long QT syndrome and a case of Andersen–Tawil syndrome. GVUS were also found in the patients with Brugada syndrome and with idiopathic ventricular fibrillation. In all these patients, GVUS were detected in genes that encode the potassium channel (KCNQ1), sodium channel (SCN5A), calcium channel (CACNA1C), ankyrin (ANK2), desmoplakin (DSP), and titin (TTN).
A comment about the limitations of genetic studiesThe need and usefulness of the genetic study has been emphasized in relatives of patients with any of these syndromes. Concomitantly with the greater availability of the molecular analysis, there is evidence that genetic studies also have certain limitations as those observed in our patients. Schwartz et al.5 clearly presented these limitations by pointing out that: only in 25 to 80% of cases (depending on the disease) the mutation could be detected, many of the variants detected might not be “pathogenic”. That is, not all can be considered as a cause of the disease, as there are very few mutations in which functional studies have been performed to confirm their pathogenicity. We want to emphasize the difficulties encountered in the interpretation of this new technique as a large number of GVUS would be identified by this method as observed in patient 8. The increasing number of screened genes would increase the number of potential pathogenic mutations and false positive tests.
We would like to point out that, until now, mainly the “exome” (exons or coding sequences) has been analyzed. To clear out the mystery of these false negative tests, a lot of recent research has been focused on the introns (non-coding sequences) but we would like to mention that studies on epigenetic modifications, which can somehow alter the genetic expression in general are also underway.
ConclusionGenetic molecular analysis performed with this NGS approach allows the identification of DNA sequence changes in 87% of cases, including pathogenic mutations in 50%. The high percentage of GVUS obtained require co-segregation and functional studies to confirm their possible pathogenic significance.
FundingNo endorsement of any kind received to conduct this study/article.
Conflict of interestThe authors declare no conflict of interest.
We would like to express our gratitude to Sistemas Genómicos (Valencia, Spain) by sponsoring the next-generation sequencing in these Mexican Patients.