Background: Lymphomas are B and/or T cell clonal neoplasms in various states of differentiation, characteristically compromising lymph nodes. They are constituted by B and T lymphocytes that reach the node by chemokine-mediated recruitment including CXCL13. Hypoxia-inducible transcription factor (HIF-1α) plays a role in cellular adaptation to oxygen concentration changes. It also regulates expression of chemokines such as CXCL12, CCL20, and CCL5 as well as some of their receptors such as CCR7 and CXCR4.
Methods: We performed in silico analysis of the CXCL13 promoter, pharmacologic modulation of HIF-1α activity and, using reporter plasmids, site-directed mutation and DNA-protein interaction analysis we analyzed the relation between HIF-1α activity and CXCL13 expression. Moreover, we did tissue microarray and immunohistochemistry to see the expression of HIF-1α and CXCL13.
Results: This study detected three possible HIF-1α binding sites suggesting that this chemokine may be regulated by the CXCL13 transcription factor. We showed that CXCL13 expression is directly dependent, whereby an increase in HIF-1α activity increases CXCL13 expression and decreased HIF-1α activity in turn decreases CXCL13 expression. We proved that HIF-1α transcriptionally regulates the expression of CXCL13 in a direct manner. We established that HIF-1α and CXCL13 are greatly overexpressed in the most aggressive pediatric lymphomas.
Conclusions: For the first time, this study showed that HIF-1α directly regulates transcriptional CXCL13 and that both proteins are overexpressed in the most aggressive forms of pediatric lymphoma. This suggests that they may play a significant role in the pathogenesis of pediatric non-Hodgkin's lymphoma.
Introduction
Non-Hodgkin's lymphomas (NHL) are among the ten most common neoplasms in pediatric populations in Mexico and worldwide. They are malignant clonal immune system malignancies resulting from the proliferation of malignantly transformed lymphoid progenitor cells in some stage of differentiation. Lymph nodes are the main tissue giving rise to most NHL.1
CXCL13 is an important chemokine in the formation of the B zone of lymph nodes by recruiting B cells to lymph node follicles and to the spleen.2 CXCL13 is known to be essential to the development of lymph nodes and Peyer's patches.3 The first observations on the possible role of CXCL13 in cancer development were obtained in gastric lymphomas associated with Helicobacter pylori infection4 and AIDS-related non-Hodgkin's lymphomas.5 Subsequent studies have confirmed that this molecule is overexpressed in different type of neoplasms such as T cell angioimmunoblastic lymphoma6 and central nervous system lymphoma,7 among others. The selective cooperation of CXCL13 and CCL19 (ligands of CXCR5 and CCR7, respectively) is reflected in apoptosis resistance of CD23+ CD5+ B cells in acute (B-ALL) and chronic (B-CLL) lymphatic leukemia.6
Little is known in regard to the transcriptional regulation of chemokines. In 2009, the CXCL13 gene was shown to be a target of the NF-κB alternative pathway.8 Using in silico analysis of the CXCL13 promoter, we found three possible hypoxia-inducible transcription factor (HIF-1α) binding sites; this transcription factor is known to be a pivotal mediator of the transcriptional response to hypoxic stress. Aside from its adaptive function in response to cellular stress, recent studies have highlighted the role of HIF-1α in physiological and pathological processes.9 Overexpression of this transcription factor is an indicator of unfavorable prognoses in several types of cancer. Also, HIF-1α regulates ∼150 genes, including those of chemokines such as CCL2, CCL12 and some of their receptors.10 Macrophages have also been reported to induce overexpression of chemokines CXCL12, CCL10, CXCL1, CCR7, CCL3 and CXCR4 in hypoxic conditions.11 To date, however, no publication has suggested that HIF-1α participates in CXCL13 regulation that, as previously mentioned, plays a pivotal role in the formation of lymph nodes, the primary affected organ in NHL. The goal of this study was to evaluate whether HIF-1α transcriptionally regulates CXCL13 and to determine if the expression of both proteins correlates with the aggressiveness of different types of pediatric NHL.
Materials and methods
Pharmacologic inhibition of HIF-1α degradation
Prostate cancer cells (PC-3) were treated with the HIF-1α degradation inhibitor ethyl-3,4-hydroxybenzoate (EDHB) or vehicle (DMSO) at different concentrations (25, 50 and 100 μM). After 24 h, cells were harvested and slides were prepared for each used concentration in order to evaluate HIF-1α and CXCL13 expression by immunohistochemical techniques.
Furthermore, PC-3 cells were treated with EDHB 25 μM and cultured for 1.5, 3, 6, 12, 18 and 24 h. Cells were harvested and stained for HIF-1α and CXCL13 with anti-HIF-1α antibodies conjugated to CSF and anti-CXCL13 antibodies conjugated to PE (R&D Systems, Minneapolis, MN) at each time interval. The expression of each protein was determined by flow cytometry after normalizing for the vehicle (DMSO).
Pharmacologic modulation of HIF-1α expression
PC-3 cells were placed in an eight-well Labtek chamber slide for cell culture (Thermo Fisher Scientific, Waltham, MA) and either treated or not with 25 μM of the HIF-1α degradation inhibitor EDHB (Sigma, St. Louis, MO), 1 μM of the HIF-1α inhibitor 2-methoxyestradiol (Sigma) or vehicle (DMSO). Cells were cultured for 24 h. The medium was removed and the cells were carefully washed in PBS and fixed with acetone. The slides were stored for a maximum of 5 days in PBS at 4 ºC before immunofluorescence staining.
Obtaining a lymphoma cell line overexpressing HIF-1α
Ramos cells (Burkitt's lymphoma) were treated in a constant manner with 25 mM EDHB and cultured for 2 months until a clone overexpressing HIF-1α was obtained; it was named RsRs. RsRs cells were maintained in culture in 25 μM EDHB.
Immunocytochemistry
Immunocytochemical techniques were used to evaluate the expression of HIF-1α and CXCL13 using specific antibodies: human anti-HIF-1α (R&D Systems) or human anti-CXCL13/ BLC/BCA-1 (R&D Systems) both at a 1:100 dilution and their respective IgG isotype control (Santa Cruz Biotechnology, Santa Cruz, CA). Briefly, after recovering the antigen from the slides, they were placed in a 0.01 M sodium citrate solution and incubated in a water bath for 20 min. They were blocked with normal pig serum for 2 h and the antibodies of interest were added. The preparations were incubated with the primary antibody overnight at 4 ºC in a humid chamber. The universal biotinylated Link (Dako, Carpinteria, CA) and streptavidin conjugated with horseradish peroxidase (HPR) (Dako) were used as secondary antibodies. Color was generated by adding the substrate diaminobenzidine (DAB) for 1 to 2 min and counterstaining was performed with hematoxylin. Finally, the cells were dehydrated and covered with resin.
Flow cytometry
The intracellular expression of HIF-1α and CXCL13 was evaluated using the Foxp3 Fixation/Permeabilization kit (eBiosciences). Briefly, 2 × 105 PC-3, Ramos and RsRs cells were fixed and permeabilized with fixating and permeabilizing regulating solution according to the manufacturer's instructions, for 1.5 h at 4 ºC in the dark. The cells were washed in regulating solution to permeabilize 1X. Staining was conducted with the antibodies: fluorescein-conjugated anti-HIF-1α and phycoerythrin-conjugated anti-CXCL13 (R&D Systems), at a 1:10 dilution. The antibodies were incubated for 1 h at 4 ºC, in the dark. The cells were washed twice with regulating solution to permeabilize 1X. They were resuspended in PBS and the median fluorescence intensity was measured in a FACS Calibur cytometer.
Cloning of the CXCL13 promoter
Based on a bioinformatic analysis of the TESS and Genomatix databases, we identified the CXCL13 promoter region. We recorded 2000 bp upstream from the transcription initiation site using the NCBI database. Specific oligonucleotides were designed for promoter amplification (997 bp). The oligonucleotide sequences were as follows: sense 5′-CGA TCT GCA GGG CAG TTT AGT GGT ACT CTA G-3′; antisense 5′-AC GGA TCC AAC TTC ATT CTG TCT GGA GGT A-3′. Human peripheral blood mononuclear cell DNA (100 ng) was used to amplify the promoter. DNA was extracted with the QIamp DNA mini kit (QIAGEN). PCR was conducted in the following conditions: 95 ºC, 3 min/35 cycles (95 ºC 30 sec; 62.3 ºC 30 sec; 72 ºC 1 min)/72 ºC 2 min. The promoter was cloned between the pLGFP-N1 (derived from pEGFP-N1) reporter plasmid sites Pst1 and BamHI. With the resultant plasmid (pCXCL13-GFP), DH5α bacteria were transformed and cultured in the presence of kanamycin (30 μg/μL); antibiotic-resistant colonies were selected for plasmid DNA extraction and corroboration by enzymatic digestion with PstI and BamHI. Purified pCXCL13-GFP plasmid DNA was used to transfect PC-3 cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The transfection and expression controls were the plasmid without a promoter (pLGFP, negative control) and the plasmid with the cytomegalovirus promoter (pCMV-EGFP, positive control).
Site-drected mutation
Three pairs of specific oligonucleotides were designed for each mutation. According to the Quick Change II XL Site Directed kit's specifications, the sequences were as follows: M1 CXCL13 Sense 5′-CTC CCT TCA TGC AGA AAA AGG CAC ACA AAG CAG GG-3′, antisense 5′-CCC TGC TTT GTG TGC CTT TTT CTG CAT GAA GGG AG-3′; M2 CXCL13 Sense 5′-TCC ACA TGC TAG GGT TTT TAT GCG CTC GAC GG-3′, antisense 5′-CCG TCG AGC GCA TAA AAA CCC TAG CAT GTG GA-3′; M3 CXCL13 Sense 5′-TGG TAC TGC AAA CCT TTT GCC TGG ACT CAG AGC-3′, antisense 5′-GCT CTG AGT CCA GGC AAA AGG TTT GCA GTA CCA-3′. The mutation was conducted with the CXCL13-GFP plasmid. PCR was performed with the reagents and in accordance with the Platinum Pfx DNA Polymerase manufacturer's instructions. PCR program used for the mutations was as follows: 95 ºC 1 min; 18 cycles (95 ºC 50 sec; 60 ºC 50 sec; 68 ºC 1 min); 68 ºC 7 min.
Sequencing
Briefly, with 400 ng plasmid DNA obtained from each mutant, PCR was conducted with Big Dye 3.1 using specific oligonucleotides to the region where the mutation occurred. The oligonucleotides used were as follows: for pM1 CXCL13 [5′-G TTT ATG TCA GAC TGT AAT ATG G-3′], for M2 CXCL13 [5′-AGA GAT GGA AAA GAC TGG TAG G-3′], for M3 CXCL13 [5′-CAA ATT TAT AAA GTT TGC AAG AGA AG-3′]. The PCR program was as follows: 96 ºC 1 min; 25 cycles (96 ºC 10 sec; 50 ºC 5 sec; 60 ºC 4 min) and 4 ºC. The sequencing reaction was performed in an ABI Prism 310 sequencer (Department of Genetics, CINVESTAV, IPN, Mexico).
Evaluation of the function of the promoter and its mutants
PC-3 cells (2 × 105) were transfected with plasmid DNA from each mutation using lipofectamine 2000. The cells were cultured for 48 h at 37 ºC and 5% CO2. Cells were subsequently harvested, fixed in formalin and expression of the GFP protein was determined by flow cytometry (FACS Calibur, Hospital de Especialidades, CMN SXXI, Mexico).
Chromatin Immunoprecipitation
PC-3 cells were harvested and fixed in 1% formaldehyde. Chromatin Immunoprecipitation (ChIP) analysis was conducted with the MAGnify Chromatin Immunoprecipitation System (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. Briefly, cells were lysed and sonicated to obtain DNA fragments between 600 and 1000 bp. The anti-HIF-1α antibody (MAB 1536, R&D Systems) or normal mouse IgG (Invitrogen, Carlsbad, CA) were bound to the protein A/G magnetic beads for 3.5 h at 4 ºC.
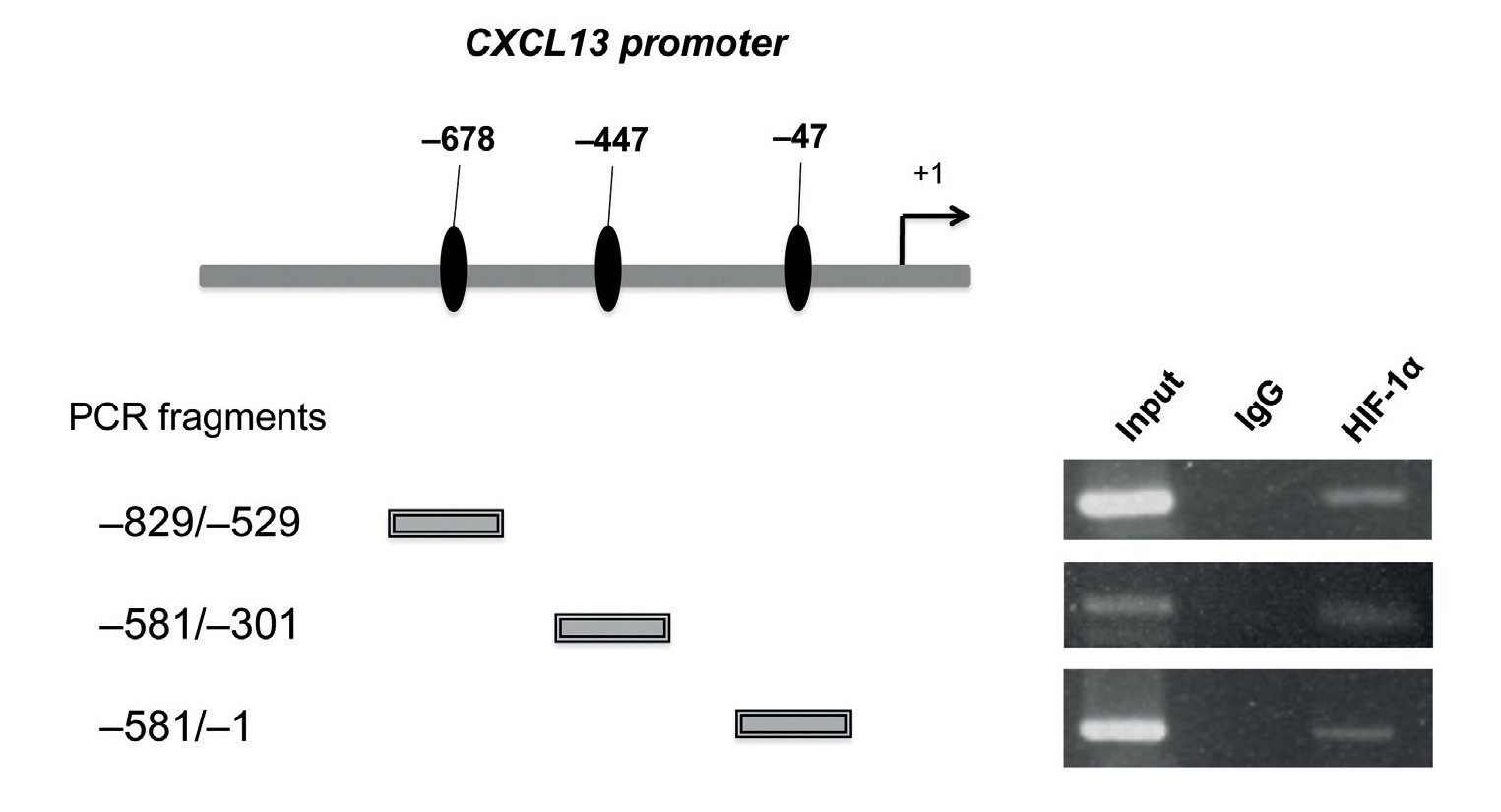
Fragmented chromatin was incubated with the anti-HIF-1α antibody (R&D Systems) or mouse IgG (Invitrogen) bound to the magnetic beads, overnight at 4 ºC. The eluted immune complexes were digested with proteinases K (20 mg/mL) and subsequently purified with magnetic beads (Invitrogen). End-point PCR was conducted to amplify the sites of interest. The oligonucleotide sequences were as follows: -678 pb: sense 5′-G TTT ATG TCA GAC TGT AAT ATG G-3′, antisense 5′-CAG ATC CTA ACG CTG ATT ATG-3′, -447 pb sense 5′-AGA GAT GGA AAA GAC TGG TAG G-3′ antisense 5′-GCT TGG AAG CTC CAA AGC AC-3′, -47 sense 5′-AGA GAT GGA AAA GAC TGG TAG G-3′ antisense 5′-CTT CAT TCT GTC TGG AGG TAG-3′.
Tissue microarray from pediatric patients with non-hodgkin's lymphoma
A tissue microarray was constructed with 58 biopsies from pediatric patients with lymphoma. Lymph node slices were embedded in paraffin. They were obtained from patients at the Hospital Infantil de México Federico Gómez and arrayed according to the Microarray technique with a semi-automatic ATA-100 Chemicon system (Chemicon's Advanced Tissue Array, Chemicon, Temecula, CA). Eleven cases were diagnosed as lymphoblastic lymphoma, 13 as Burkitt's lymphoma, six as large B cell lymphoma and four as anaplastic lymphoma.
Immunohistochemistry
Serial slices of the microarray were deparaffinized and hydrated. Antigen retrieval was performed using sodium citrate (0.01 M, pH 6.0). Endogenous peroxidase activity was inhibited with methanol and 3% hydrogen peroxide. Antibody-tissue non-immune binding was blocked by immersion in universal blocking solution and 1% bovine serum albumin for 60 min. Tissue slices were incubated overnight at room temperature with anti-HIF-1α (MAB1536, R&D Systems) and anti-CXCL13 antibodies (AF-801, R&D Systems). After washing, slides were incubated with the universal Link biotinylated antibody (Dako) and then with streptavidin conjugated with horseradish peroxidase (HPR); color was generated by adding DAB (diaminobenzidine). The reaction was stopped and samples were counterstained with hematoxylin. Tissues were dehydrated and the preparations were covered with resin and dried at room temperature.
Expression analysis of each microarray protein was determined by digital pathology with a ScanScope CS digital processor (Aperio, San Diego, CA).
Results
Modulation of HIF-1α expression in PC-3 cells affects the expression of CXCL13
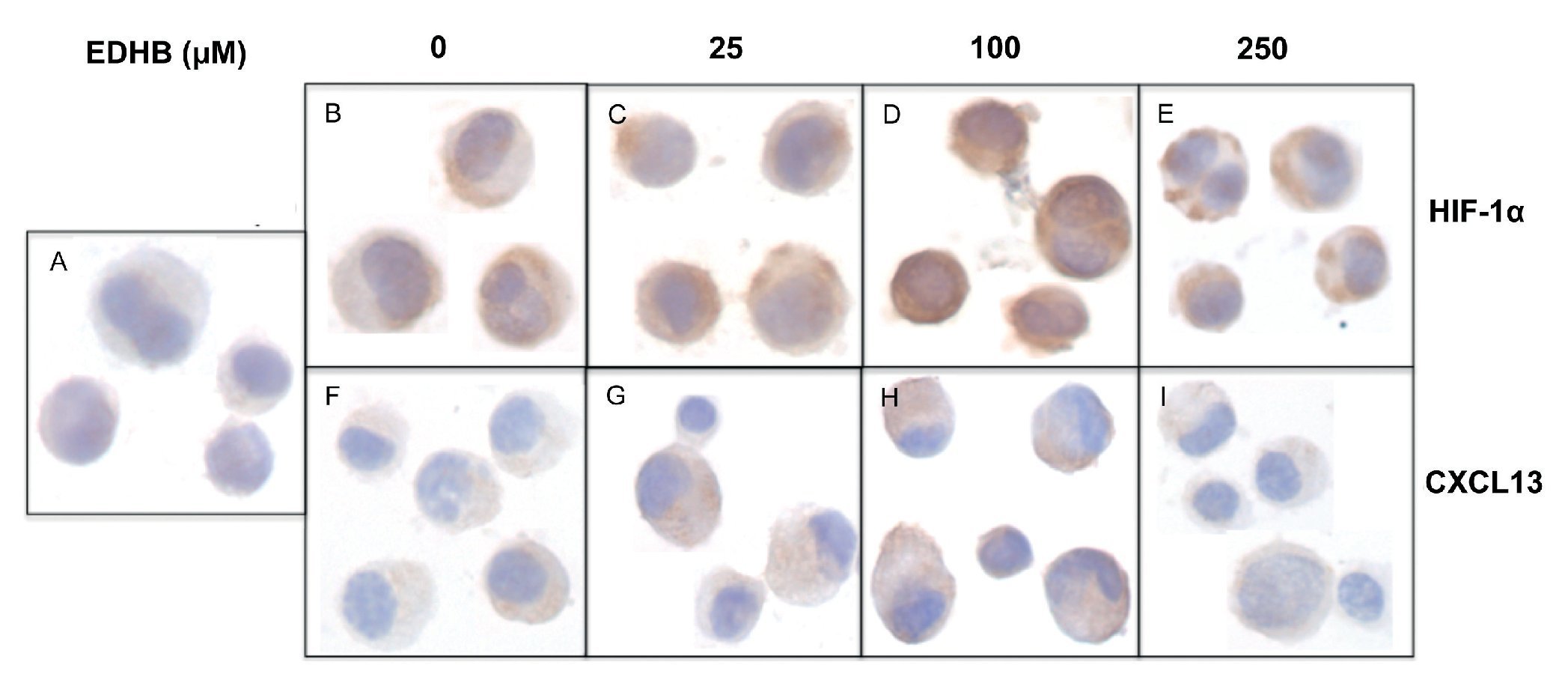
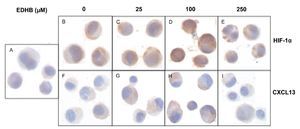
To evaluate the effect of an increase in HIF-1α on CXCL13 expression, PC-3 cells were treated with the HIF-1α degradation inhibitor EDHB at various concentrations (25, 100 and 250 μM) for 24 h. Using immunocytochemistry and with specific antibodies, the expression of both proteins was determined. Results showed that the accumulation of HIF-1α (Figs. 1B-1E) did affect CXCL13 expression, significantly increasing when cells were treated with 25 and 100 μM EDHB in comparison with untreated cells. These results suggest that CXCL13 expression increases in direct proportion to the accumulation of HIF-1α. However, expression of both proteins decreases at a concentration of 250 μM EDHB. This decrease may be due to cell death as suggested by the cellular morphology on immunostaining.
Figure 1 The induction of HIF-1α accumulation increases CXCL13 expression in PC-3 cells. PC-3 cells were treated for 24 h with different concentrations (25, 100 and 250 μM) of the HIF-1α degradation inhibitor (EDHB). HIF-1α and CXCL13 expression was evaluated by immunocytochemistry using specific anti-HIF-1α and anti-CXCL13 antibodies. A) isotype control (cells treated with DMSO); B) and F) untreated cells; C) and G) cells treated with 25 μM EDHB; D) and H) cells treated with 100 mM EDHB; E) and I) cells treated with 250 μM EDHB. (Magnification ×100).
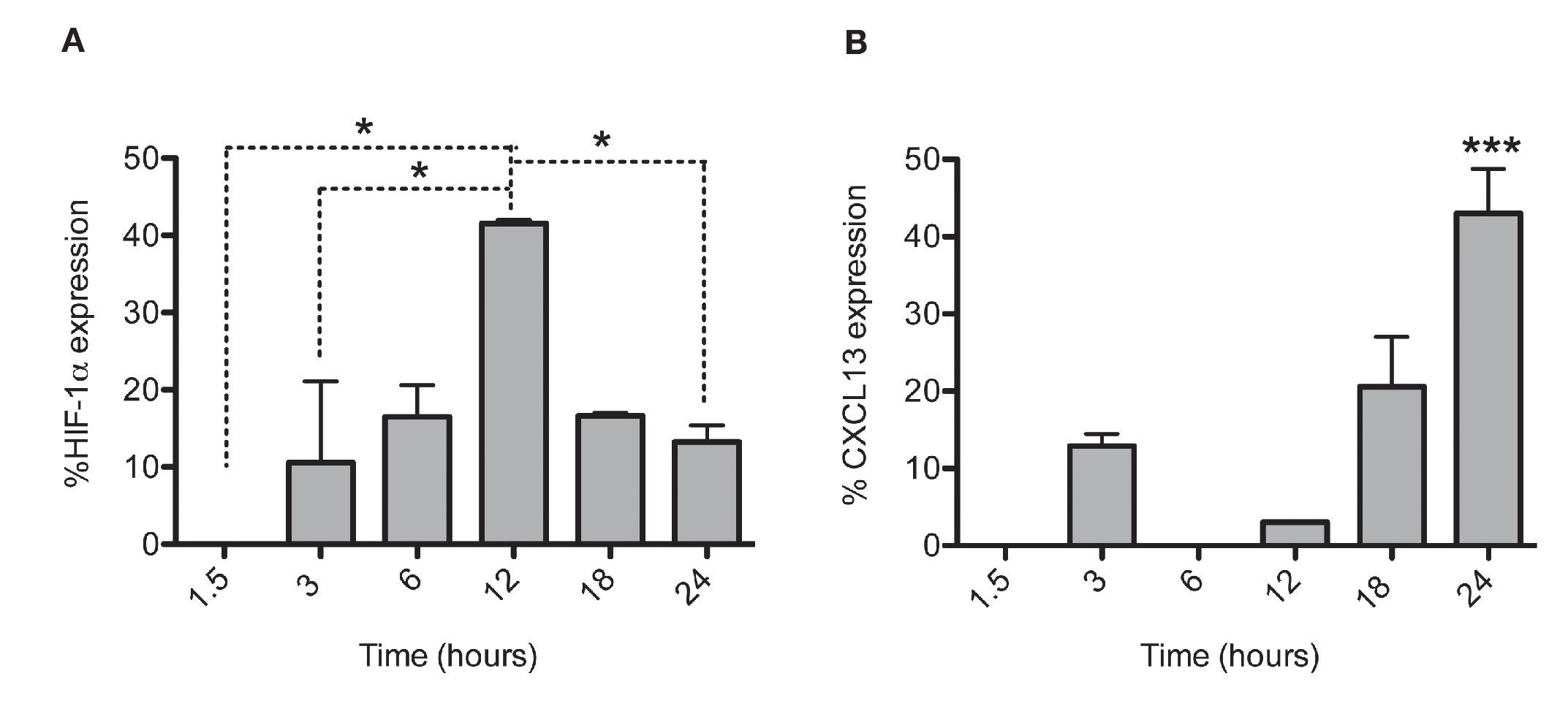
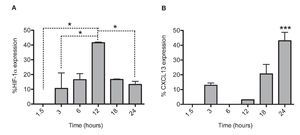
After establishing the optimal concentrations with which EDHB-mediated HIF-1α accumulation induces CXCL13 expression, we evaluated the optimal duration of EDHB treatment to induce it. We conducted a time kinetics protocol whereby PC-3 cells were treated with 25 μM EDHB at different time points (1.5, 3, 6, 12, 18 and 24 h). They were subsequently immunostained to determine HIF-1α and CXCL13 expression at different time points by flow cytometry (Figs. 2A and 2B, respectively). For each EDHB time point, a corresponding control was assigned (DMSO). We considered 100% as the mean fluorescence intensity of the cells incubated only with the vehicle. Figure 2A shows the results obtained for HIF-1α total expression that significantly increased after 12 h of treatment. However, at that point, CXCL13 expression was minimal (Figure 2B) but increased significantly after 18 h and was most evident after 24 h. At this point, HIF-1α expression began to decline.
Figure 2 HIF-1α accumulation induces CXCL13 expression over time. The expression of each protein was quantified by flow cytometry. Expression was normalized according to the median fluorescence intensity with the drug's vehicle (DMSO). Average and standard deviation results of three independent experiments. *p = 0.01 ***p = 0.0001 (one-way ANOVA).
HIF-1α-mediated CXCL13 expression regulation
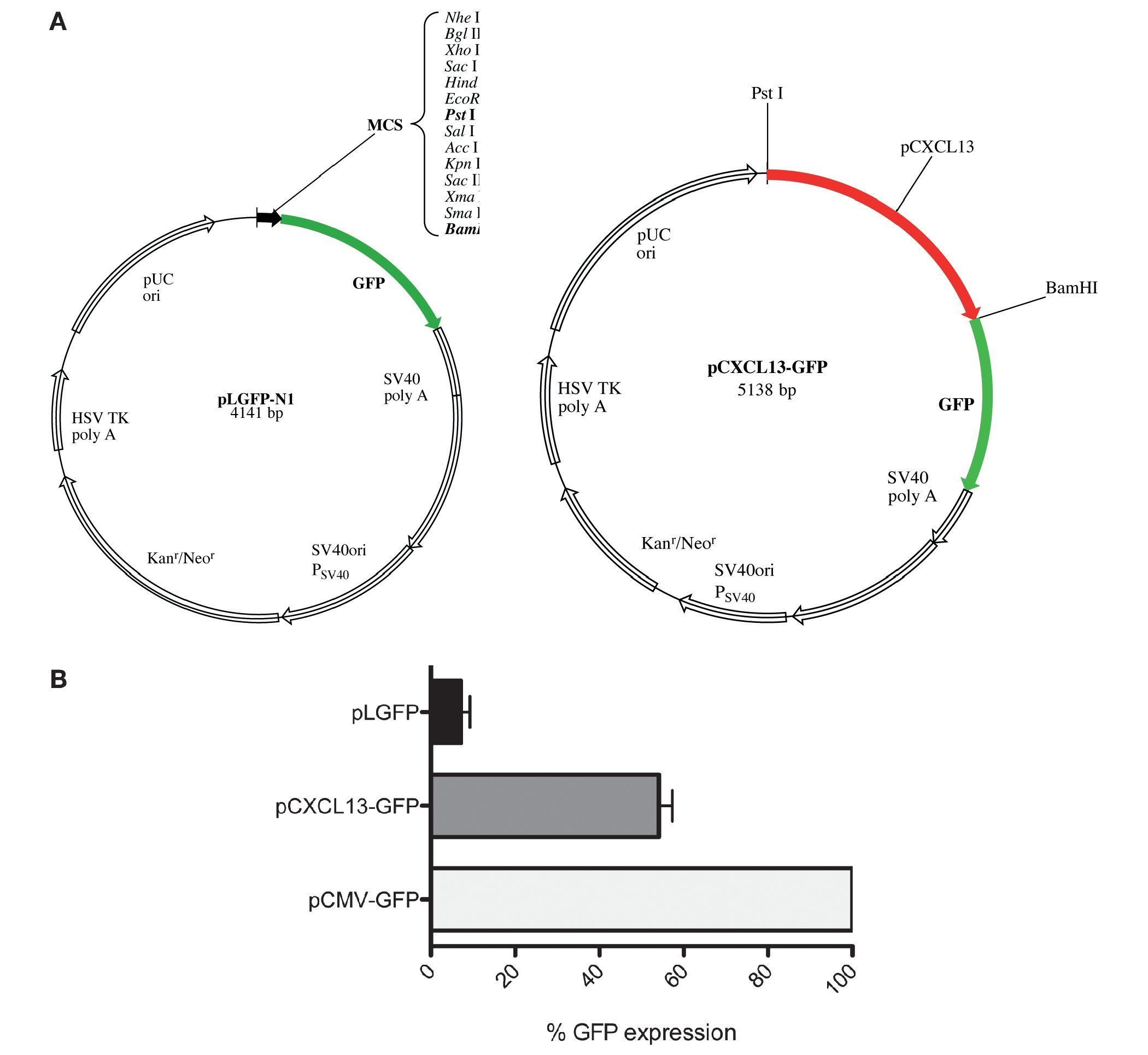
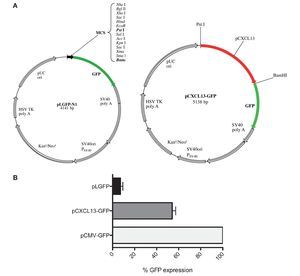
After evaluating the direct effect of HIF-1α expression modulation on CXCL13 expression and based on the bioinformatics results suggesting that CXCL13 has three putative HIF-1α binding sites (-47, -447 and -678 bp), we considered the possibility that this transcription factor may play an active role in CXCL13 transcriptional regulation. In order to evaluate this, we used reporter plasmid technology and cloned the CXCL13 promoter in pLGFP-N1. We obtained a construct named pCXCL13-GFP composed of 5138 bp (Fig. 3A).
Figure 3 GFP protein activity with CXCL13 promoter. (A) pLGFP-N1 plasmid map into which CXCL13 promoter was inserted. (B) Evaluation of the promoter's functionality. PC-3 cells were transfected with lipofectamine with each construct and median fluorescence intensity of the reporter protein (GFP) was quantified by flow cytometry (FACS, Calibur). Transfection efficiency was ∼30%.
Evaluation of the CXCL13 promoter's functionality was performed by analysis of GFP reporter protein expression (pCXCL13-GFP) by flow cytometry. Figure 3B shows the obtained results in terms of GFP mean fluorescence intensity. As expected, expression of this protein was increased in cells transfected with the positive control, pCMV-GFP. The CXCL13 promoter was functional as reflected by GFP expression in cells transfected with the plasmid containing this chemokine's promoter (pCXCL13-GFP).
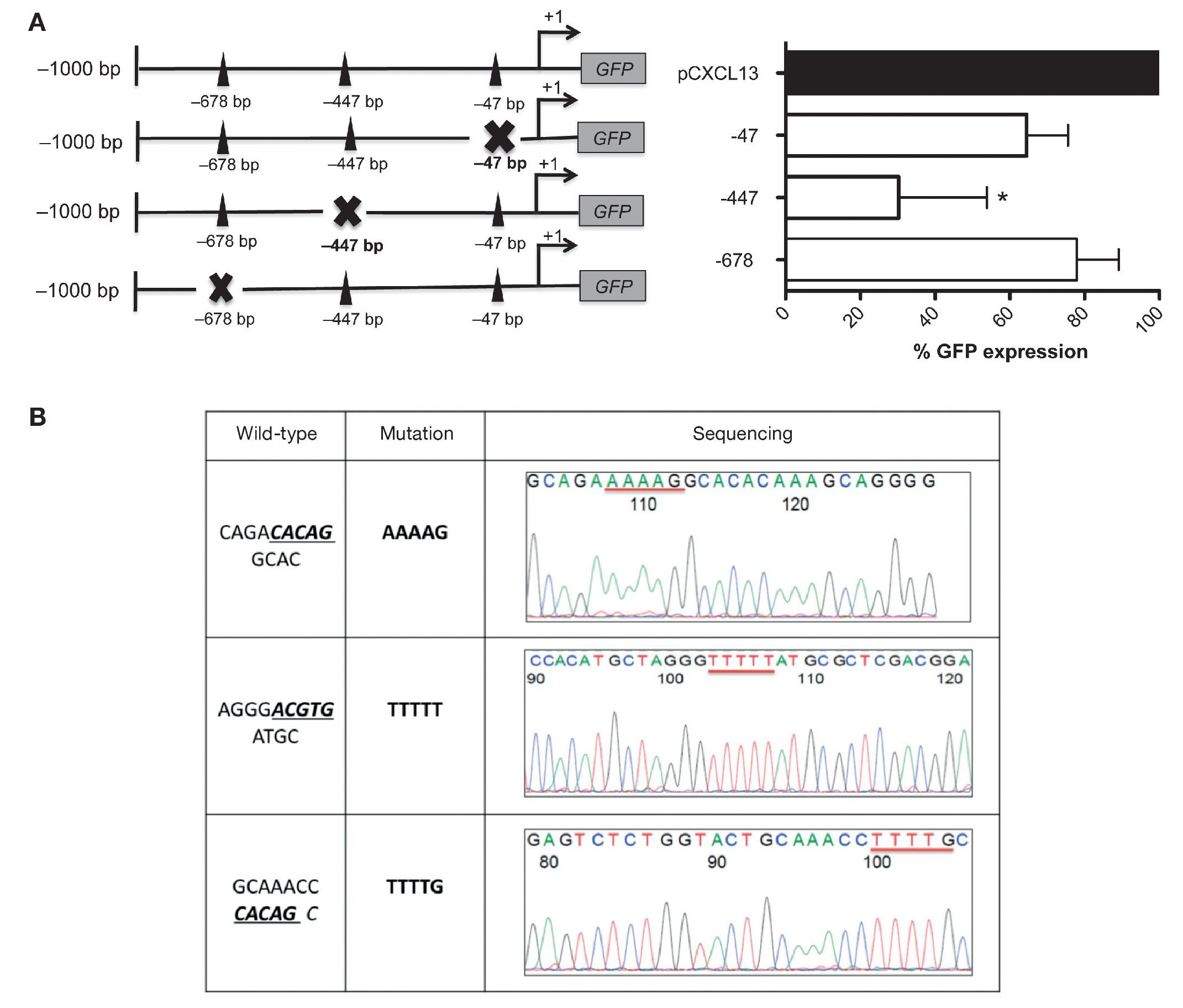
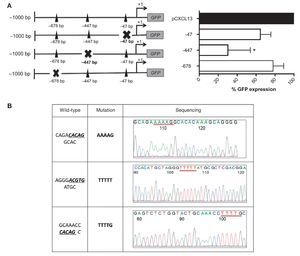
These results allowed us to continue the evaluation of the relevance of each HIF-1α binding site in the regulation of the CXCL13 promoter; we conducted site-directed mutagenesis at each possible site (Figure 4A). Subsequently, the mutation sequence of the CXCL13 promoter was verified by sequencing as described in detail in Materials and Methods. The histograms in Figure 4B show 100% homology. Figure 4A shows GFP expression in the cells transfected with the plasmids containing each HIF-1α mutation site. GFP expression is significantly decreased in the cells transfected with the plasmid containing the -447 site mutation when compared with that in cells transfected with the plasmid containing the entire promoter (pCXCL13). Cells transfected with the plasmids containing the -47 and -678 mutations show a tendency to display a decrease in expression in comparison to cells transfected with the entire plasmid. This decrease, however, was not statistically significant.
Figure 4 Mutations in the different HIF-1α sites decreased CXCL13 promoter activity. (A) Sketch of the CXCL13 promoter and possible HIF-1α-promoter binding sites. GFP reporter protein expression at each directed site mutation in pCXCL13-GFP and each possible HIF-1α-CXCL13 promoter binding site. PC-3 cells were transfected with the different constructs and promoter activity was evaluated according to the GFP reporter protein's median fluorescence intensity by flow cytometry. The average of three independent experiments is shown. ANOVA *p <0.05. (B) Sequencing electropherograms of each mutation conducted in the pCXCL13-GFP plasmid.
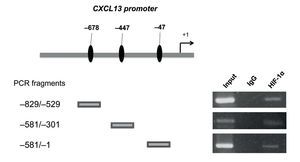
After determining that HIF-1α plays an important role in regulating CXCL13 expression, it was of interest to prove whether this transcription factor regulates the chemokine's expression at the transcription level by directly binding to its promoter. We thus conducted ChIP analysis. We initially designed specific oligonucleotides for each possible HIF-1α binding site of the CXCL13 promoter in accordance with the bioinformatics analysis. In order to identify the fragments of immunoprecipitated DNA with the anti-HIF-1α antibody, we standardized the PCR for each site using PC-3 cell genomic DNA. We obtained fragments of 587 bp (site -47), 280 bp (site -447) and 320 bp (site -678). The PC-3 cell fragmented chromatin was used in the ChIP assay along with a specific HIF-1α antibody and PCR was conducted for each possible binding site. Figure 5 shows that HIF-1α is capable of interacting with the CXCL13 promoter in the three sites detected by bioinformatics analysis. These results reflect the fact that HIF-1α positively upregulates CXCL13 expression by directly binding to any of the three sites on this chemokine's promoter. However, the results of the site-directed mutation indicated that the site -447 bp mutation showed the greatest relevance in the chemokine's expression. The Input (unfragmented chromatin) was used as a positive control in the PCR for each site of interest.
Figure 5 HIF-1α may interact with all three CXCL13 promoter binding sites. Chromatin immunoprecipitation (ChIP) was conducted in PC-3 cells with the anti-HIF-1α antibody. Interaction sites were identified by PCR using specific oligonucleotides for each possible HIF-1α binding site in the CXCL13 promoter.
HIF-1α-mediated CXCL13 expression in non-Hodgkin's lymphomas cells
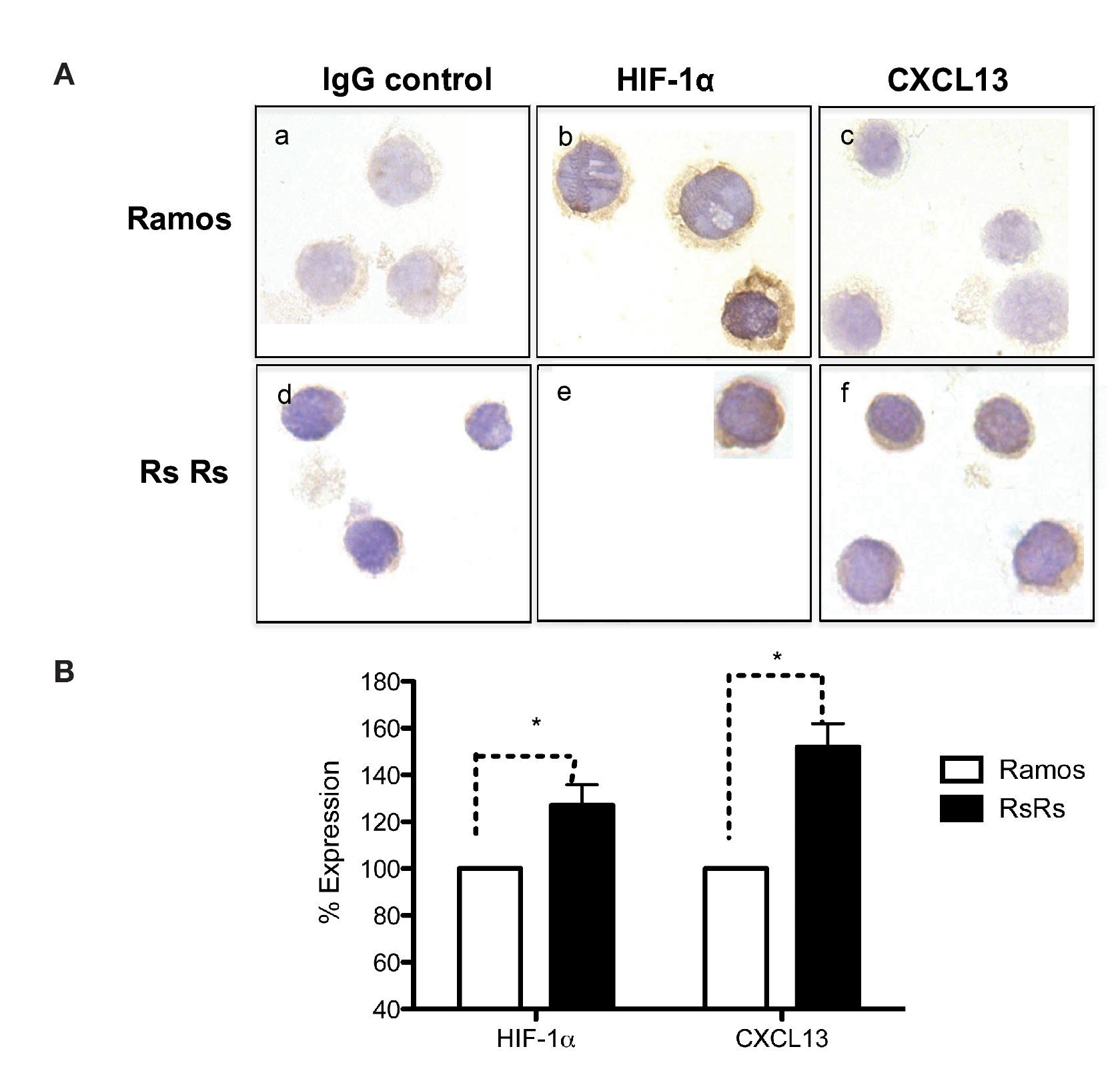
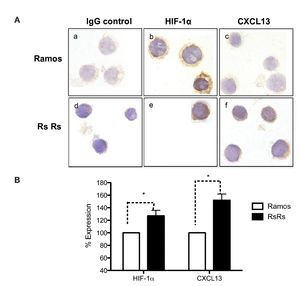
As previously mentioned, it is known that CXCL13 plays an important role in the formation of lymph nodes and that it is highly expressed in different types of lymphomas. We were therefore interested in evaluating whether HIF-1α-mediated regulation of CXCL13 expression could be biologically relevant in NHL cells. The expression of both proteins was evaluated in Ramos cells (Burkitt's lymphoma) that constitutively express HIF-1α as well as in RsRs cells derived from Ramos cell line. These Ramos cells, dubbed RsRs, were treated for a long time period with the HIF-1α degradation inhibitor EDHB; therefore, they mostly express this transcription regulator. Figure 6A shows the immunocytochemistry results in which cells expressing high HIF-1α levels (RsRs) also highly express CXCL13 when compared with Ramos cells. The expression of both proteins was also evaluated by flow cytometry (Fig. 6B). Results were consistent with HIF-1α overexpression and its direct effect on CXCL13 expression. Our results strongly suggest that CXCL13 is regulated by HIF-1α in NHL cells.
Figure 6 Induction of HIF-1α accumulation in lymphoma cells increases CXCL13 expression. (A) Expression was evaluated by immunocytochemistry with specific HIF-1α y CXCL13 antibodies. a) and d) isotype controls; b) and e) HIF-1α expression; c) and f) CXCL13 expression. (B) Flow cytometry. Ramos and RsRs cells were stained intracellularly to determine HIF-1α and CXCL13 expression using specific antibodies bound to a fluorochrome. *p <0.05
HIF-1α and CXCL13 expression in pediatric patient non-hodgkin's lymphomas histological subtypes
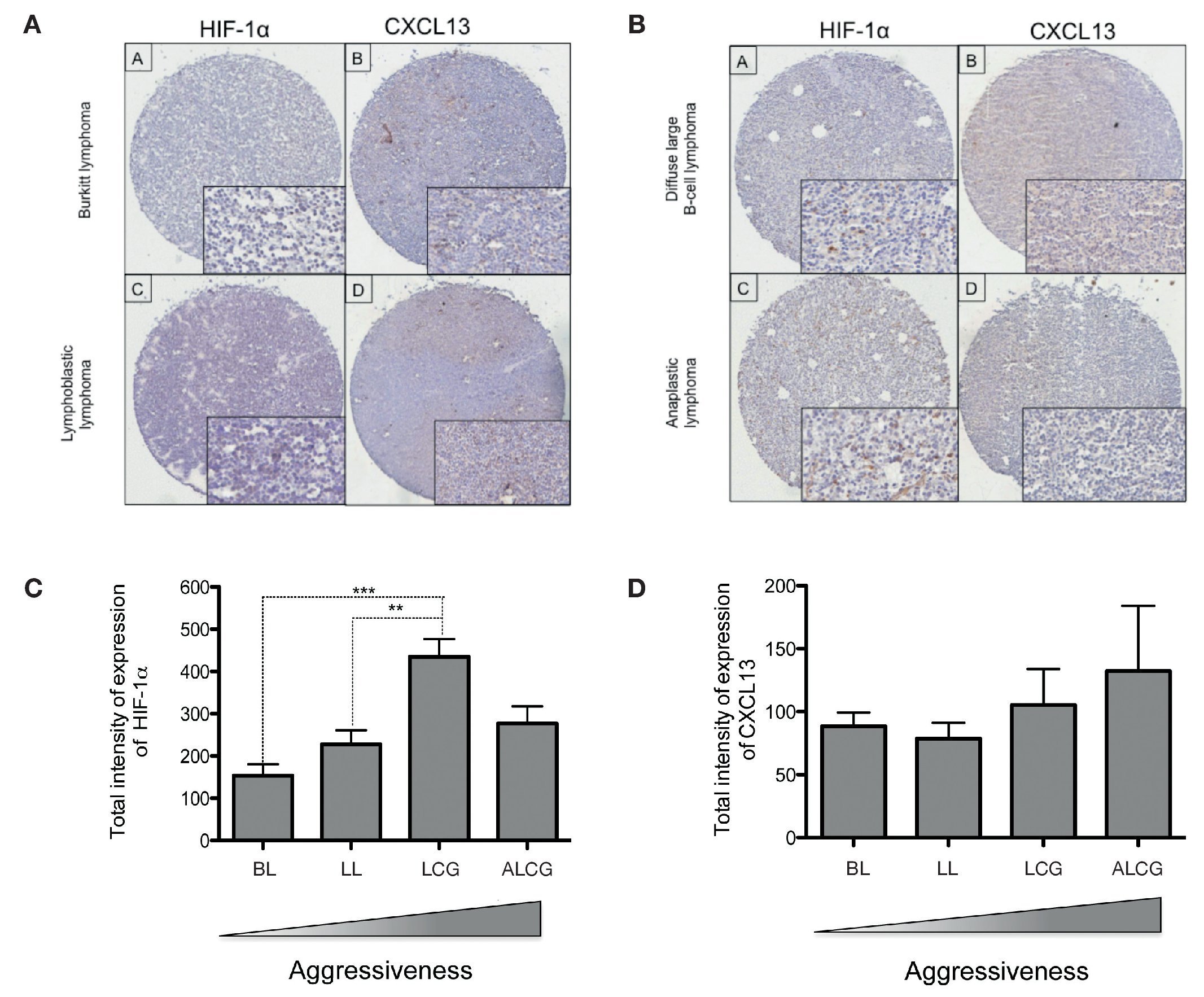
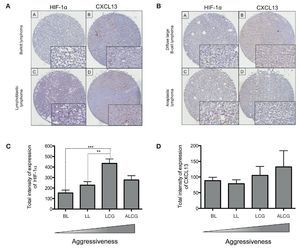
In order to determine if CXCL13 transcription regulation by HIF-1α may be of biological relevance in patients with NHL, we conducted expression analysis of both proteins in biopsies obtained from pediatric patients with this disease. HIF-1α and CXCL13 expression were evaluated by immunohistochemistry in a tissue microarray obtained from pediatric patient biopsies and included four NHL subtypes. Figure 7A is a representative microphotograph of the least aggressive histological subtypes (Burkitt's and lymphoblastic lymphomas) and shows that expression of both proteins is lower than that in more aggressive subtypes (anaplastic and large B cell lymphomas) (Fig. 7B). Quantification of the expression of both proteins was conducted with a digital pathology system and is shown in Figures 7C and 7D. HIF-1α expression increases significantly in large B cell lymphoma when compared to with the less aggressive subtypes (Fig. 7C), whereas no significant difference in CXCL13 expression was detected (Fig. 7D). A possible correlation between HIF-1α and CXCL13 expression according to the type of lymphoma was also evaluated; Spearman r correlation factor of 0.2 was found in the least aggressive lymphomas.
Figure 7 HIF-1α and CXCL13 expression are increased in the most aggressive NHL. Proteins HIF-1α and CXCL13 were identified by immunohistochemistry with specific antibodies. (A) Microphotograpic representation of Burkitt's lymphoma (BL) and lymphoblastic lymphoma (LL). (B) Large B cell lymphoma (LCG) and anaplastic lymphoma (ALCG). Quantification of total HIF-1α and CXCL13 expression intensity in each NHL subtype was analyzed using the automated image digitizer, ScanScope CS. (C) HIF-1α expression in different pediatric NHL. ANOVA ** p <0.05, ***p <0.0001, respectively. (D) CXCL13 expression in different types of pediatric NHL.
Discussion
This study has shown that with pharmacological modulation, an increase in HIF-1α activity increased CXCL13 expression and, conversely, decreased HIF-1α activity decreases expression of the chemokine. Moreover, using reporter plasmids, site-directed mutation of the CXCL13 promoter and DNA-protein interaction analysis we demonstrated that HIF-1α directly transcriptionally regulates CXCL13. Interestingly, using tissue microarray and immunohistochemistry techniques, we showed that HIF-1α and CXCL13 expression are significantly increased in the most aggressive pediatric lymphomas.
In this study, EDBH and 2-Me were used as pharmacological modulators of HIF-1α activity and expression in order to evaluate this transcription factor's role in CXCL13 expression. Results showed that treatment of PC-3 cells with EDHB at different time points has an effect on CXCL13 expression. An accumulation of HIF-1α was observed as well as an increase in CXCL13 expression after treatment with 25 and 100 μM EDHB when compared with control cells treated only with the vehicle. However, at a concentration of 250 μM, the expression of both proteins decreased in comparison with the control. This decreased expression was associated with a cell death phenotype suggesting that high EDHB concentrations are toxic to cells. We thus decided to use a concentration of 25 μM in subsequent experiments that did show a significant increase in HIF-1α but cell viability was not affected. Using time kinetics, we found that the highest HIF-1α expression in PC-3 cells induced by 25 μM EDHB was after 12 h but was not reflected by an increase in CXCL13 expression. This suggests that HIF-1α accumulates in the cytoplasm at earlier time points (12 h) but that longer time periods are required (18 and 24 h) for HIF-1α to be translocated to the nucleus and bind to the hypoxia response elements in the target gene's promoter, in this case, CXCL13.
Little is known in regard to CXCL13 transcription regulation. There are studies showing that CXCL13 is an NF-κB target gene, which may lead us to think that HIF-1α's modulatory effect on CXCL13 expression may indirectly reflect the activity of the alternative NF-κB pathway.8 In order to demonstrate the direct participation of HIF-1α in CXCL13 expression, site-directed mutation assays with a reporter plasmid and chromatin immunoprecipitation (ChIP) were performed. We observed that the plasmid reporter assays and site mutations showed that the -447 site (-ACGTG—the most studied consensus site to date) was indeed the most important one in the induction of CXCL13 expression. However, ChIP assays revealed that this transcription factor binds to three putative sites (-47, -447, -678). This proves that HIF-1α plays a pivotal and direct role in the transcription regulation of CXCL13.
CXCL13 is an important chemokine in the formation of the nodal B zone that attracts B lymphocytes expressing the chemokine's only receptor, CXCR5. In the tumor micro-environment of B cell neoplasms, different stromal cell populations have been shown to favor malignant cell survival including several subgroups of T cells that promote disease progression, T cells with anti-tumor effects and endothelial cells that organize neoangiogenesis.12 Hence, chemokines such as CXCL13 and its receptor CXCR5 may play a role in lymphomagenesis of B cells. Also, little is known about the role of HIF-1α and CXCL13 in NHL. Some studies have been conducted in adult patients,13 but there are no studies in the pediatric population in whom NHL is the fifth or sixth most common cause of cancer, depending on the geographical area studied. We therefore evaluated HIF-1α and CXCL13 expression in vitro in a Burkitt lymphoma cell line (Ramos cells). Ramos cells did indeed increase their HIF-1α expression after exposure to EDHB (RsRs). Protein expression was evaluated by immunocytochemistry and flow cytometry. Results again proved that CXCL13 regulation mediated by HIF-1α also occurs in NHL cells. This may be of relevance in the formation of lymph nodes.14
We therefore studied the expression of these proteins in biopsies obtained from NHL pediatric patients. HIF-1α and CXCL13 expression was determined in 35 pediatric NHL patients by immunohistochemistry in tissue microarrays and the analysis and quantification of their expression was performed with an automated image analyzer. The most aggressive lymphoma subtypes overexpressed HIF-1α when compared with the less aggressive subtypes. This difference was statistically significant. Although there is a tendency for CXCL13 to be more expressed in the more aggressive subtypes, the difference is not statistically significant.
These results suggest, for the first time, that HIF-1α is overexpressed in one of the most aggressive pediatric lymphoma subtypes and that it may play a role in the pathogenesis of the disease.
This is the first report demonstrating that the hypoxiainducible factor participates in CXCL13 transcription regulation as well as the relevance of these two proteins in pediatric NHL.
Funding
This work was supported by Mexico's Federal Funds Grant HIM/2009/009 (SHY), Mexican Foundation of Health and Merck Grant F769 (SHY), and Master scholarship degrees grant (KMM) CONACyT-Becaria and Miembros del Patronato del HIM (SHY).
Conflict of interest
The authors declare no conflict of interest of any nature.
Received 8 November 2013;
accepted 12 December 2013
* Corresponding author.
E-mail address:shuertay@yahoo.com (S. Huerta-Yepez).