Breast cancer is one of the most common malignancies affecting women. Recent investigations have revealed a major role of ion channels in cancer. The transient receptor potential melastatin-2 (TRPM2) is a plasma membrane and lysosomal channel with important roles in cell migration and cell death in immune cells and tumor cells.
MethodsIn this study, we investigated the prognostic value of TRPM2 channel in breast cancer, analyzing public databases compiled in Oncomine™ (Thermo Fisher, Ann Arbor, MI) and online Kaplan-Meier Plotter platforms.
ResultsThe results revealed that TRPM2 mRNA overexpression is significant in situ and invasive breast carcinoma compared to normal breast tissue. Furthermore, multi-gene validation using Oncomine™ showed that this channel is coexpressed with proteins related to cellular migration, transformation, and apoptosis. On the other hand, Kaplan-Meier analysis exhibited that low expression of TRPM2 could be used to predict poor outcome in ER- and HER2+ breast carcinoma patients.
ConclusionsTRPM2 is a promising biomarker for aggressiveness of breast cancer, and a potential target for the development of new therapies.
El cáncer de mama es la neoplasia maligna más común que afecta a mujeres. Estudios recientes han revelado un papel importante de los canales iónicos en el cáncer. El receptor de potencial transitorio melastatin-2 (TRPM2) es un canal que se expresa en la membrana plasmática y en los lisosomas; posee funciones importantes en la migración y muerte celular de células inmunes y tumorales.
MétodosEn este estudio se investigó el valor pronóstico del canal TRPM2 en cáncer mama. Se realizó el análisis de bases de datos públicos empleando las plataformas OncomineTM (Thermo Fisher, Ann Arbor, MI) y Kaplan-Meier Plotter.
ResultadosLos resultados mostraron que el mRNA de TRPM2 se sobreexpresa significativamente en los carcinomas de mama in situ e invasivo en comparación con el tejido mamario normal. Además, la validación de múltiples genes empleando OncomineTM reveló que este canal se coexpresa con proteínas relacionadas con la migración celular, la transformación celular y apoptosis. Por otra lado, el análisis de la sobrevivencia promedio usando curvas Kaplan-Meier mostró que la baja expresión de TRPM2 podría utilizarse como un marcador de pronóstico pobre en pacientes con carcinoma de mama receptor de estrógeno negativo (ER-) y receptor 2 del factor de crecimiento epidermal positivo (HER2+).
ConclusionesEl TRPM2 podría emplearse como biomarcador de agresividad en cáncer de mama, y como blanco para el desarrollo de nuevas terapias.
Breast cancer is the most common malignancy among women, and the first cause of death in this population worldwide (Cancer Today: http://gco.iarc.fr/today/home). These tumors are highly heterogeneous, and although tremendous advances have been made in their prevention, diagnosis, and treatment, there is a continuous search for new biomarkers to identify new subtypes and to predict their clinical behaviors and response to treatment.1–3 Breast cancer is subtyped into four groups according to the expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor 2 (HER2). These groups are ER-positive or PR-positive/HER2-negative (ER+ or PR+/HER2-); ER+ or PR+/HER2+; ER-/PR-/HER2+ (known as HER2+); and ER-/PR-/HER2- (triple negative).1–3 Most breast cancers are ER+, which are well differentiated and less aggressive tumors, and have a better prognosis than tumors with few or no estrogen receptors (ER-/PR-tumors).1–3 Besides, ER expression has been used as an indicator of endocrine responsiveness although only 50% of ER+ breast tumors respond to anti-estrogen or aromatase inhibitors. In addition, a small number of ER- tumors can respond to hormonal therapy.1–3 Moreover, PR+ tumors (65-75% of all breast cancers) are hardly ER-. Consequently, PR has been used to discard false ER negativity that would exclude some patients from receiving endocrine therapy. ER+PR- tumors show lower mortality rate and are less responsive to endocrine treatment than ER+PR+. In contrast, ER-PR- tumors have higher recurrence rate, lower survival and do not respond to endocrine treatment.1–3
On the other hand, 13-20% of invasive ductal breast cancers are HER2+, which spread more quickly than other subtypes. Likewise, tumor suppressor TP53, which plays a role in cell proliferation, survival, apoptosis and genomic integrity, has been used as prognostic marker independent of tumor size, node status, and hormone receptor expression. Approximately 25–30% of breast tumors have a mutation on TP53.1,4 Furthermore, TP53-PR- tumors have the worse prognosis among all breast cancers.1,4 Breast cancer can also be classified based on the status of ER, HER2, and the proliferation marker KI67 into five intrinsic subtypes: basal (ER-/HER2-), luminal A (ER+/HER2-/KI67 low), luminal B (ER+/HER2-/KI67 high and ER+/HER2+), HER 2+ (HER2+/ER1-) and normal-like tumors.1–3 Microscopically, they are also grouped into grades (1-3; less to more aggressive), indicating the speed by which the tumor will grow and extend.1–3
Recently, many studies have investigated the role of ion channels in several types of cancer, including breast cancer.5–7 For instance, members of the Transient Receptor Potential (TRP) channel family, TRPC1, TRPC6, and TRPM7 have been associated with cell proliferation in breast cancer.7 However, studies on the utility of these proteins as biomarkers are still scarce. In the present study, we investigated the prognostic value of the TRPM2 channel —a TRP family member involved in cell migration and cell death8—in breast cancer, analyzing public databases. We found consistently higher expression of TRPM2 in invasive breast tumors in comparison to in situ tumors and normal tissue, making it a promising biomarker for aggressiveness and a potential target for the development of new therapies.
2Methods2.1Oncomine™ Platform analysisThe expression of TRPM2 gene in breast cancer was analyzed and visualized using the Oncomine™ Platform (Thermo Fisher, Ann Arbor, MI; http://www.oncomine.org).9 For this, we compared TRPM2 mRNA from clinical specimens of cancer versus normal patient datasets with more than 151 samples. We selected p<0.01 as a threshold, 1.5-fold change and gene rank in top 10% because these values are analytical and statistically significant. We analyzed the results for their p-values, fold changes, and cancer subtypes.
2.2Kaplan-Meier Plotter analysisThe prognostic value of TRPM2 gene in breast cancer was analyzed using Kaplan-Meier Plotter (http://kmplot.com/analysis), a database that combines gene expression and clinical data.10,11 To date, Kaplan-Meier Plotter contains information of 54,675 genes and their effect on survival in 4,142 breast cancer patients with a mean follow-up of 69, 40, 49 and 33 months. We focused our analysis on overall survival and relapse-free survival patient information with a 10-year follow-up. The samples were divided into two groups (high and low TRPM2 expression levels) using the median gene expression value and were compared using a Kaplan-Meier survival plot. The hazard ratio with 95% confidence intervals and log Rank p-value were calculated and displayed. We used the best specific probes (JetSet probes) that recognized TRPM2.
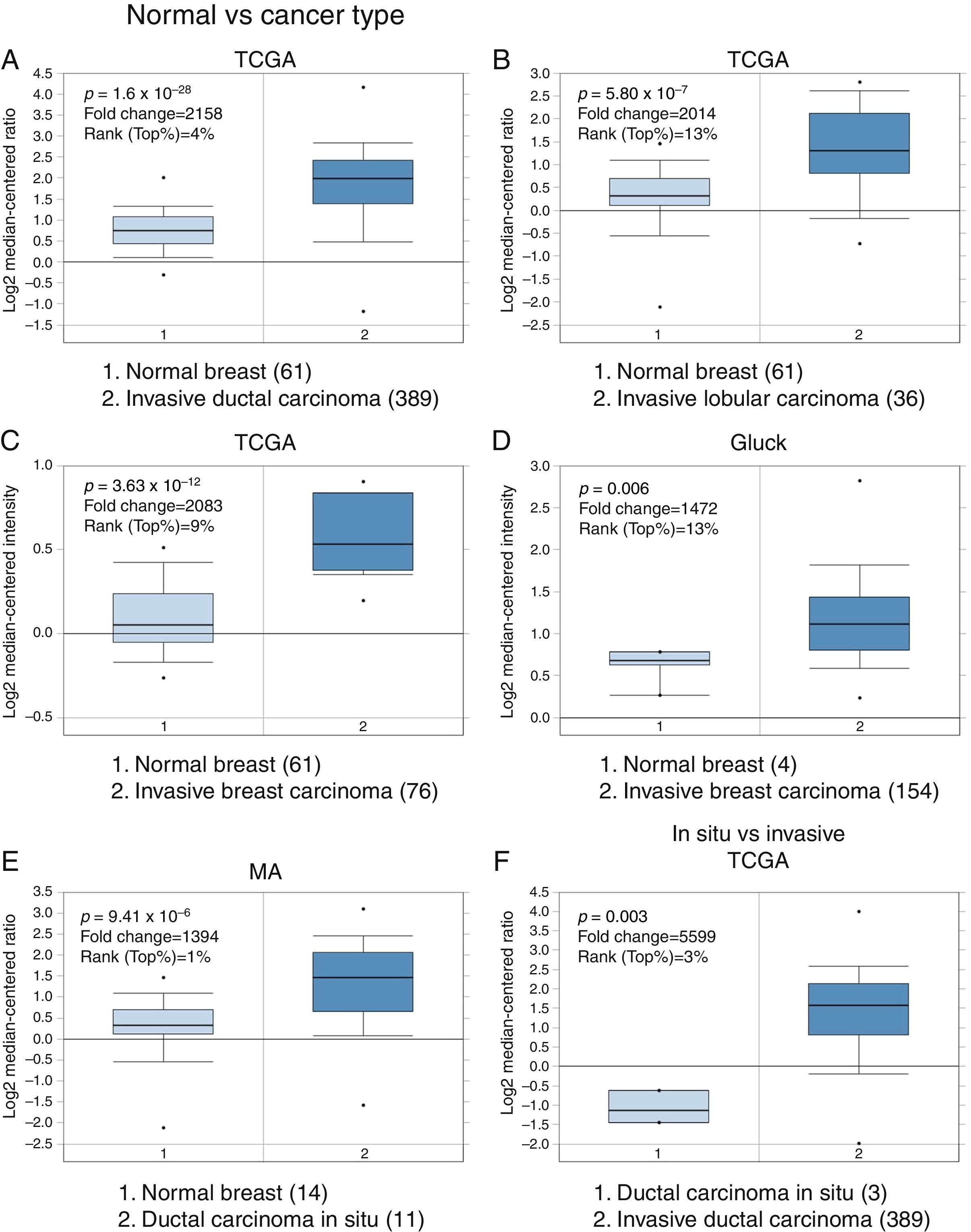
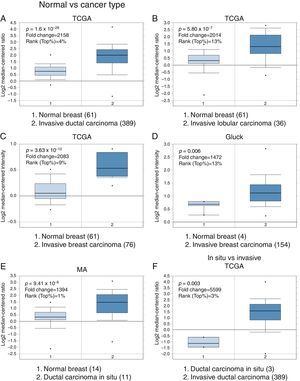
3Results3.1Differential expression of the TRPM2 transcript in normal and malignant breast tissueTo our knowledge, no data have been published previously on TRPM2 expression level or its possible prognostic value in breast cancer. Therefore, we decided to perform data mining on mRNA expression profiles on normal and malignant breast cancer sets in the Oncomine™ Platform, as previously described. Three independent datasets deposited in Oncomine™ were selected to identify the differential mRNA expression of TRPM2 channel associated with breast cancer. Oncomine™-The Cancer Genome Atlas (TCGA) interactive interface showed 501 different tissue samples of breast carcinoma and 61 of normal material. As shown in Figure 1, TRPM2 was significantly overexpressed in 389 invasive ductal breast carcinomas (IDC) compared to 61 normal breast material (fold change=2.158, Figure 1A). Afterward, we performed the same analysis on the invasive lobular carcinoma (ILC) samples from the TCGA dataset. We found that TRPM2 expression was higher in ILC when compared to normal breast tissue (fold change=2.014, Figure 1B), as well as in invasive breast carcinoma (fold change=2.083, Figure 1C), which comprises a mixture of cancerous ductal and lobular cells. The second dataset in Oncomine™, Gluck dataset, also reflected the same transcriptional features for TRPM2 in 154 invasive breast carcinomas (fold change=1.476, p=0.006; Figure 1D) compared to normal, even though there were only four samples for normal tissue. Moreover, the analysis of the third dataset (MA dataset) showed that this channel was highly expressed in 11 samples of ductal carcinoma in situ as well. Although the sample sizes for normal and ductal carcinoma in situ were small, the p-value was significant (p=9.41×10−6). Consequently, we next compared the performance of TRPM2 channel in invasive versus in situ ductal carcinoma using the TCGA dataset. Whereas the database had only three in situ samples, we observed almost a six-fold overexpression of this gene in invasive carcinoma (fold change=5.599, Figure 1F). A box-plot of all this data, which compared medians, also showed the data to be significant. Altogether, these analyses suggest that the increase in TRPM2 expression is associated with cell transformation, particularly to invasive carcinoma. Therefore, this protein is a significant predictor of tumor invasion.
Boxplot results of a meta-analysis of TRPM2 expression in normal breast and invasive carcinoma. The Oncomine™ dataset was reviewed according to the differential analysis order by over-expression in distinct microarrays. (A) In TCGA dataset, TRPM2 was overexpressed in a subset of invasive ductal carcinoma compared with normal breast tissue (p=1.69e-28) and (B) in invasive lobular carcinoma (p=5.80e-7) and showed significantly higher expression in (C) invasive breast carcinoma (p=3.63e-10). (D) In Gluck database, TRPM2 was overexpressed in invasive breast carcinoma (p=0.006). (E) TRPM2 was highly expressed in eleven samples of ductal carcinoma in situ from MA dataset (p=9.41e-6). (F) TRPM2 expression was significantly higher in invasive ductal carcinoma than in ductal carcinoma in situ, suggesting that the use of this channel as a biomarker should be investigated. The number of samples in each subtype is shown in parentheses.
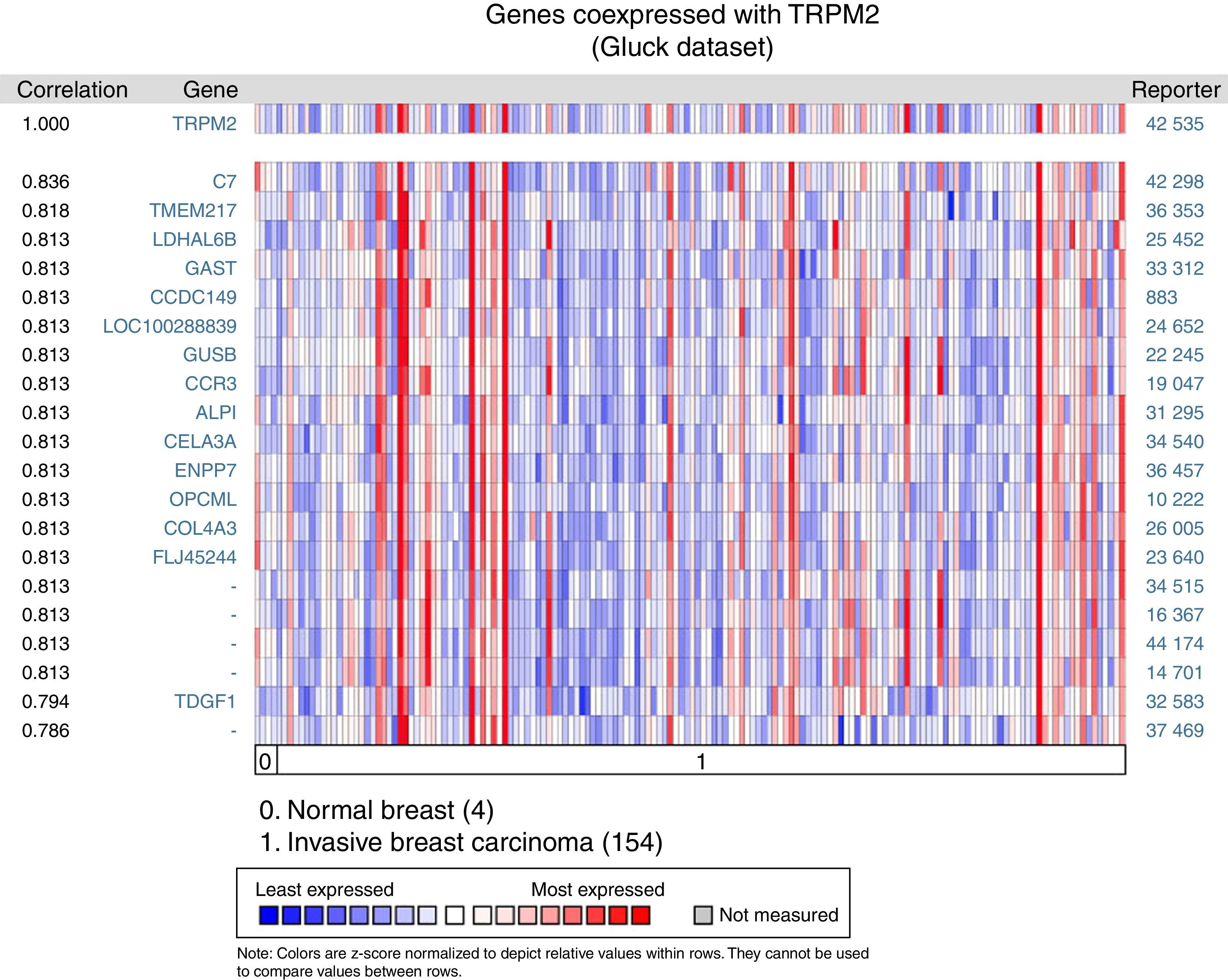
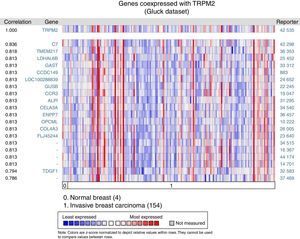
Given that cell migration requires more than one element, we figured out that TRPM2 channel is not the only one gene that is highly expressed in invasive breast carcinoma. We performed a multi-gene validation using Oncomine™-Gluck dataset to point out the highest correlation value ∼1, to spot those genes coordinately expressed with TRPM2. This validation used 154 invasive breast carcinoma and four normal samples.
The matching gene expression pattern filtered 15 characterized genes [C7, complement component 7; TMEM217, transmembrane protein 217; LDHAL6B, lactate dehydrogenase A-like 6B; GAST, gastrin; CCDC149, Coiled-coil domain-containing 149; LOC100288839, hypothetical protein LOC100288839; GUSB, glucuronidase beta; CCR3, C-C motif chemokine receptor 3; ALPI, intestinal alkaline phosphatase; CELA3A, chymotrypsin-like elastase family member 3A; ENPP7, ectonucleotide pyrophosphatase/phosphodiesterase 7; OPCML, opioid binding protein/cell adhesion molecule-like; COL4A3, collagen type IV alpha 3 (Goodpasture antigen); FLJ45244, hypothetical locus FLJ45244; TDGF1, teratocarcinoma-derived growth factor 1]. There were also five uncharacterized genes ranked in the top 10% of the dataset (Figure 2). Interestingly, since CCR3 was recently associated with breast cancer subtype,12 this data raised the possibility of studying the combined effect of TRPM2 and CCR3 overexpression in the same sample. The remaining genes were examined, and the result did not show any biological process over-represented. Intriguingly, these proteins are related to innate and adaptive immune response (C7, ALPI, OPCML), connected to cancer processes (LDHAL6B, GAST, TDGF1), and some, like ENPP7, prevent tumorigenesis. Moreover, these genes are reviewed in the literature as a route or function-related to cellular migration, transformation or apoptosis.
Biological associations of TRPM2 by meta-signature of invasive breast carcinoma. We employed the web-based Oncomine™-Gluck microarray database to analyze biological associations of high mRNA expression in invasive breast cancer. We compared cancer tissue (154 samples) to normal tissue (4 samples) and set a correlation for this screening. Heat map of gene expression across from red stands for most expressed, yellow for marginal and blue for least expressed. Twenty genes are overexpressed in invasive cancer compared with normal tissue counterpart signatures.
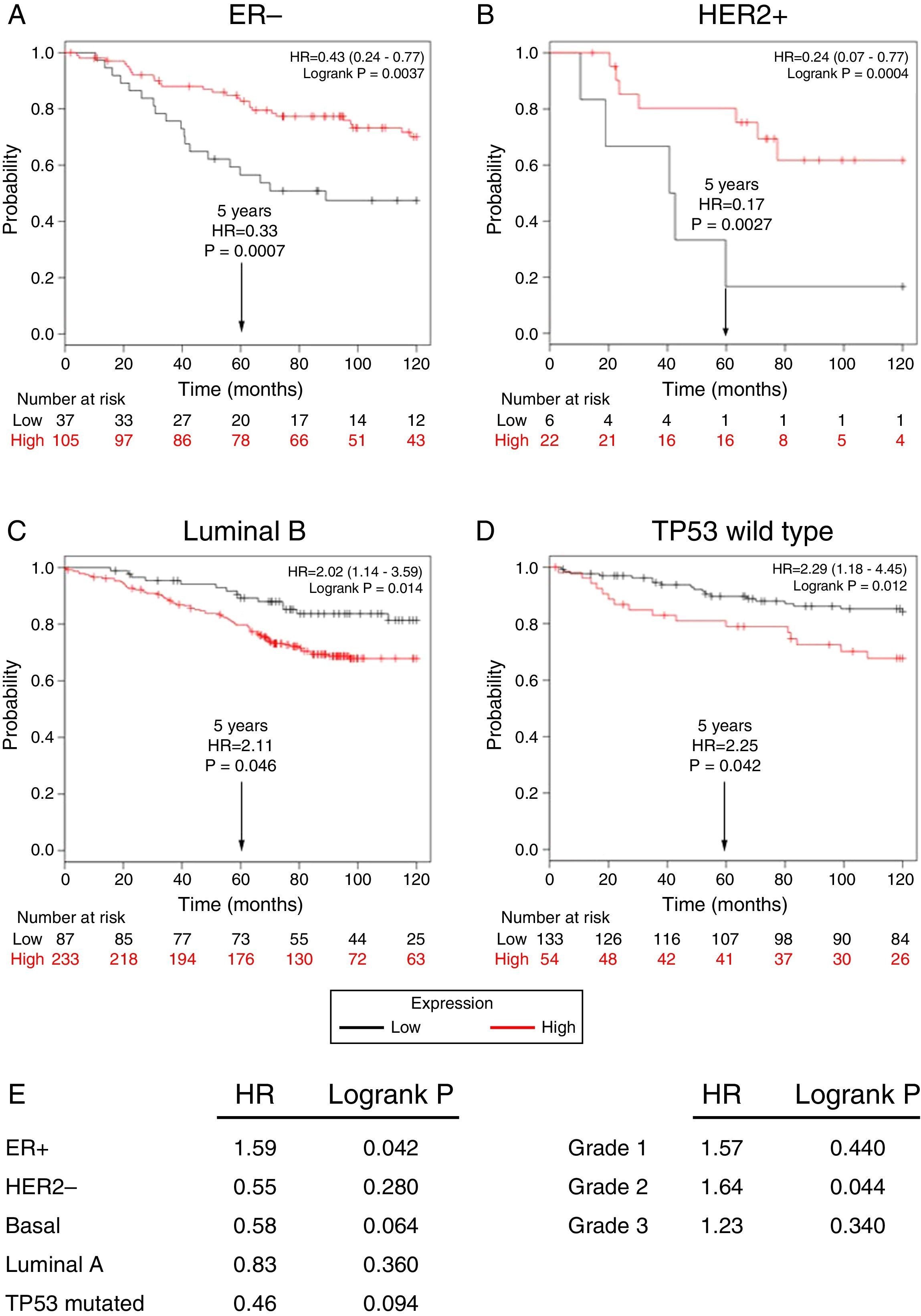
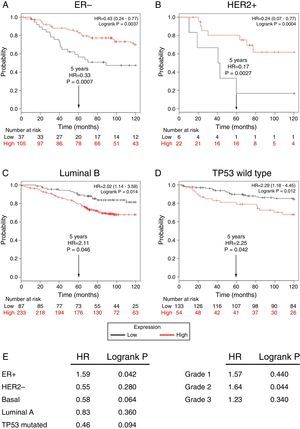
Subsequently, to establish the prognostic value of a TRPM2 expression in breast cancer overall survival and relapse-free survival, we created Kaplan-Meier plots of mRNA using data from Gene Expression Omnibus (GEO; Affymetrix microarrays only), European Generic Association (EGA) and TCGA compiled in an online plotter. Indeed, the Kaplan-Meier curves of overall survival of ER- (Figure 3A) and HER2+ (Figure 3B) patients with a 10-year follow-up, showed a correlation between low expression of TRPM2 and lower survival rates. In contrast, poor patient outcome was associated with high expression of TRPM2 in the subtypes Luminal B (Figure 3C), TP53 wild type (Figure 3D), ER+ and grade 2 (Figure 3E) tumors. Nevertheless, p values were less significant than ER- and HER2+ subtypes (Figure 3). TRPM2 expression level had no influence on overall survival of patients with HER2-, Luminal A, mutated TP53, and tumors of grade 1 and 3 (Figure 3E). Together, these data suggest that mRNA expression of TRPM2 as a predictor of outcome may be valuable in ER- and HER2+ breast carcinomas (p=0.0037 and p=0.0094, respectively).
Low expression of TRPM2 mRNA correlates with poor outcome in patients with ER- and HER2+ breast cancer. Kaplan-Meier graphs show the overall survival prognosis of breast cancer patients censored at 120 months, based on high or low TRPM2 tumor mRNA expression. Patients with expression above the median are shown in red, and patients with expression below the median in black. (A) ER- breast cancer patients. Upon dividing the patients by TRPM2 mRNA level, those with low expression (n=37) had lower overall survival probability over time (logrank P=0.0037) than those with high expression (n=105). (B) HER2+ breast cancer patients. Patients with low expression (n=6) had lower overall survival probability over time (logrank P=0.0094) than those with high expression (n=22). Curves were statistically significant. (C) Conversely, Luminal B breast cancer patients with high expression (n=233) had lower overall survival probability over time (logrank P=0.014) than those with high expression (n=87), as well as (D) patients with TP53 wild-type breast cancer. Low expression (n=133), high expression (n=54), logrank P=0.012. Curves were statistically significant. (E) Summary of hazard ratios (HR) and logrank P values of overall survival curves of patients with ER+, HER2-, Basal, Luminal A, and TP53 mutated breast cancer subtypes, and grade 1-3 tumors.
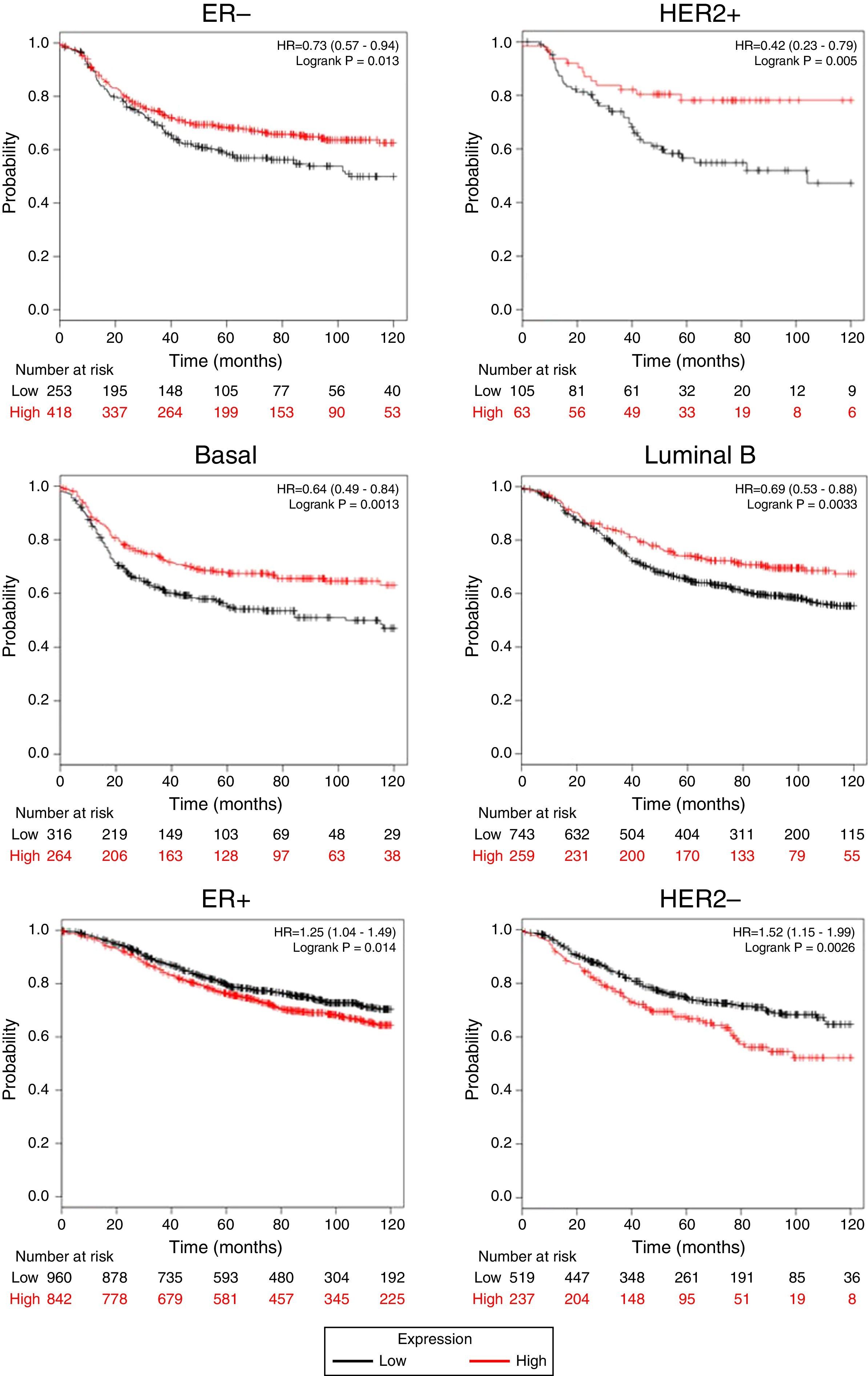
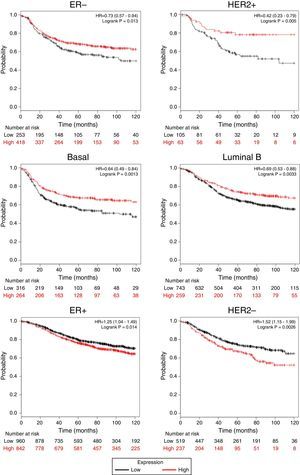
Afterward, we analyzed the impact of TRPM2 mRNA level in relapse-free survival. Figure 4 provides statistically significant evidence that the group of patients with low expression of TRPM2 in ER-, HER2+, Basal and Luminal B breast cancer presented a shorter relapse-free survival time than the panel of patients with high expression of TRPM2. On the other hand, ER+ and HER2- patients with high TRPM2 expression also presented shorter relapse-free survival time than the group with a low expression of TRPM2, although there was a lower significant difference. In addition, no significant difference was observed between both groups in TP53 wild-type, TP53 mutated, luminal A subtypes and grades 1-3 (data not shown).
TRPM2 mRNA expression correlates with relapse-free survival in patients with breast cancer. Kaplan-Meier graphs show the relapse-free survival prognosis of breast cancer patients censored at 120 months, based on high or low TRPM2 tumor mRNA expression. Red, patients with expression above the median; black, patients with expression below the median. After splitting the patients by TRPM2 mRNA level, those with low expression had a lower probability of being relapse-free over time when presented ER-, HER2+, Basal or Luminal B subtype. In contrast, high TRPM2 expression patients of ER+ and HER2- breast cancer presented slightly less relapse over time in comparison to their counterpart with low expression.
In general, these results suggest that the high expression of TRPM2 may be used as a biomarker for invasive breast carcinoma. Conversely, the low expression of TRPM2 may predict poor outcome and a higher probability of relapse in ER- and HER2+ breast carcinoma patients during treatment. Although the results showed significant differences in overall survival and relapse-free survival, p-values were higher in other subtypes; therefore, preliminary and further analysis of more public data should be performed to establish the utility of TRPM2 expression as a biomarker in those cases.
4DiscussionRecent studies have investigated the role of TRP channels in breast cancer.5–7 For instance, TRPC1, TRPC6, and TRPM7 have been associated with cell proliferation in these tumors.7 However, few investigations have been conducted on other TRP members, such as TRPM2,6 which are involved in cell migration and cell death, critical cellular processes affected in cancer cells.8 In our analysis, using Oncomine™, we found that TRPM2 is highly expressed in breast carcinoma in situ in comparison to normal breast tissue, and even expresses nearly six times more in invasive breast carcinoma samples compared to the in situ carcinoma material. These results strongly suggest that TRPM2 could be used as a molecular biomarker of tumor invasion in breast cancer. Accordingly, multi-gene validation using the Oncomine™-Gluck dataset revealed that TRPM2 co-expressed with proteins involved in cell migration (CCR3), cancer processes (LDHAL6B, GAST, TDGF1), and in preventing tumorigenesis in the intestinal tract (ENPP7). The biological relevance of these findings is still unknown. However, a recent study by Gong DH et al. has demonstrated that higher mRNA expression of CCR3 indicates a reduced risk of cancer relapse in luminal-like disease but not in triple-negative breast cancer (TNBC) and HER2- enriched cancers.12 Additionally, in human prostate cancer, the expression of the CCR3 receptor is associated with the occurrence of aggressive disease with widespread local dissemination.13 On the other hand, TRPM2 has been involved in chemokine- and bacterial peptide- activated directional migration and oxidative stress-induced cell death in several immune cells.8 Furthermore, recent data analysis of TRP channels in cancer showed that TRPM2 is overexpressed in the bladder, head and neck, liver, and lung cancer (adenocarcinoma).5 TRPM2 was also found to play a role in prostate cancer and melanoma.14,15 Besides, TRPM2 channel silencing in breast carcinoma cell lines decreased proliferation.6
Our Kaplan-Meier analysis showed that low TRPM2 could be used to predict poor outcome and a higher probability of relapse in ER- and HER2+ breast carcinoma patients during treatment. Findings of this study suggest that TRPM2 might be a potential biomarker and target for new therapies for cancer. Further analysis of more public data should be performed to establish the utility of TPRM2 as a biomarker in breast cancer subtypes.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that they have followed the protocols of their work center on the publication of patient data.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
FundingThe “Program of Improvement for the Professorship” (PRODEP), Secretariat of Public Education (Project DSA/103.5/14/10595), supported this study.
Conflict of interestThe authors declare no conflicts of interest of any nature.