An actinobacterial strain VL-RK_09 having potential antimicrobial activities was isolated from a mango orchard in Krishna District, Andhra Pradesh (India) and was identified as Arthrobacter kerguelensis. The strain A. kerguelensis VL-RK_09 exhibited a broad spectrum of in vitro antimicrobial activity against bacteria and fungi. Production of bioactive metabolites by the strain was the highest in modified yeast extract malt extract dextrose broth, as compared to other media tested. Lactose (1%) and peptone (0.5%) were found to be the most suitable carbon and nitrogen sources, respectively, for the optimum production of the bioactive metabolites. The maximum production of the bioactive metabolites was detected in the culture medium with an initial pH of 7, in which the strain was incubated for five days at 30°C under shaking conditions. Screening of secondary metabolites obtained from the culture broth led to the isolation of a compound active against a wide variety of Gram-positive and negative bacteria and fungi. The structure of the first active fraction was elucidated using Fourier transform infrared spectroscopy, electrospray ionization mass spectrometry, 1H and 13C nuclear magnetic resonance spectroscopy. The compound was identified as S,S-dipropyl carbonodithioate. This study is the first report of the occurrence of this compound in the genus Arthrobacter.
The natural products of microbial origin appear to be the most promising source of future antibiotics.1,2 A search for new antibiotics or new microbial strains producing antibiotics continues to be of utmost importance in research programs around the world because of the increases in resistant pathogens and toxicity of some known antibiotics. Nevertheless, there is an alarming scarcity of new antibiotics in the pharmaceutical industry. Therefore, to ensure the availability of effective drugs in the future, it is necessary to improve the antimicrobial use patterns and to devise strategies for the identification of new antibiotics through previously unexplored targets.3,4 Development of novel drugs with a broad-spectrum mode of action against pathogens is the need of the hour.
Actinobacteria are aerobic, Gram-positive and filamentous in nature, which form asexual spores and mycelia and have a high guanine and cytosine (GC content) in the DNA. They are the most economically and biotechnologically valuable prokaryotes with the unparalleled ability to produce bioactive secondary metabolites, notably, antibiotics, antitumor agents, immunosuppressive agents, and enzymes. In view of their excellent track record in this regard, extensive attempts have been made in the past 50 years to isolate actinobacteria from terrestrial sources for drug screening. In the present days, an exploration of rare actinobacteria for bioactive metabolite production has dramatically increased, particularly due to their unique and diverse metabolic pathways against life-threatening resistant pathogens.5 A relatively low occurrence of non-streptomycete species as producers of secondary metabolites is due to the absence of novel techniques for the isolation of these strains from the environment. Another problem is their complicated preservation and cultivation methods, which frequently require specific and unusual conditions. These are the main reasons why these microbes are considered rare organisms and why investigation and manufacturing of their products are difficult. Most isolates recovered on agar plates by conventional isolation techniques have been identified as species of Streptomyces, which are the dominant actinobacteria present in soil.2,6–8
Rare actinobacteria are widely distributed in terrestrial and marine ecosystems having different environmental conditions such as soil type, pH, humus content and humic acid characteristics of the soil.9 They are diverse and belong to many suborders, including Micrococcineae and to a variety of genera.10,11 The best-known genus of the family Micrococcaceae is Arthrobacter, which includes aerobic, catalase-positive, cocci or coryneform bacteria with lysine in their cell walls. Arthrobacter spp. exhibit great metabolic versatility and are able to produce a number of valuable substances, such as amino acids, vitamins, enzymes, specific growth factors, and polysaccharides.12 However these organisms are not well explored as secondary metabolite producers, although there have been reports of production of bioactive compounds by some species.13–16 While rare actinobacteria may provide increased chances of discovering novel structures, their genetics and physiology are poorly understood.
As a part of our ongoing research on rare actinobacteria, a promising strain VL-RK_09, with a good antimicrobial potential was identified as Arthrobacter kerguelensis. The strain was isolated from a soil sample collected at a mango orchard in Vissannapet, Krishna District, Andhra Pradesh (India), and its DNA sequence was deposited to Genbank under accession number KJ787652. Since the components of culture media and cultural conditions often influence the production of bioactive metabolites by microorganisms.17–21 An attempt was made to enhance the production of bioactive metabolites and to characterize the metabolites of A. kerguelensis VL-RK_09.
Materials and methodsChemicalsAll solvents, reagents, and media components used in this study were of extra-pure grade and procured from Merck (Mumbai, India).
Strain isolationThe strain, A. kerguelensis VL-RK_09, was isolated on yeast extract-malt extract-dextrose (YMD) agar by a soil dilution technique from a soil sample collected at a mango orchard in Vissannapet, Andhra Pradesh, India. The medium consisted of malt extract (1%), yeast extract (0.4%), dextrose (0.4%), CaCO3 (0.2%), and agar (2.0%), pH 7.0±0.2. The strain was stored on YMD agar medium at 4°C.22
Antimicrobial profile of bioactive metabolites produced by the strainTo study the antimicrobial profile, a three-day-old culture of the strain was cultivated in YMD broth (seed medium) at 30°C for 24h. Of the seed culture, 10% was transferred to the same medium for mass fermentation. The fermentation process was allowed to continue at 30°C for eight days. The production of bioactive metabolites was determined by measuring diameters of inhibition zones against test microbes. Culture filtrates harvested at regular intervals were extracted with ethyl acetate and evaporated to dry under vacuum at 35°C. The residue thus obtained was dissolved in dimethyl sulfoxide (DMSO) and tested for antimicrobial activity against test bacteria such as Staphylococcus aureus (MTCC 3160), Bacillus megaterium (NCIM 2187), Shigella flexneri (MTCC 1457), Xanthomonas campestris (MTCC 2286), Proteus vulgaris (ATCC 6380), Pseudomonas aeruginosa (ATCC 9027) and Escherichia coli (ATCC 35218) and fungus Candida albicans (MTCC 183) by an agar diffusion assay.23
Media optimizationAttempts were made to enhance the antimicrobial activity by optimizing the culture conditions such as pH, temperature, carbon sources, nitrogen sources and minerals. The bioactive metabolite production was determined after five days of incubation. Fermentation was carried out in 100mL of media in 250-mL Erlenmeyer flasks with constant shaking at 180rpm. The effect of initial pH on bioactive metabolite production by the strain was determined by adjusting pH of the production medium in the range from 4 to 10. The optimal pH achieved at this step was used for further study.24 Similarly, the optimum temperature was determined by incubating the strain at temperatures ranging from 20 to 40°C, while maintaining all other conditions at optimum levels.25 Carbon sources such as maltose, lactose, fructose, galactose, sucrose, glucose, starch, cellulose, mannitol, and sorbitol were supplemented separately at a 1% concentration into the fermentation medium.26 The effect of varying concentrations (0.5–4%) of the best carbon source on bioactive metabolite production was also investigated. Similarly, various nitrogen sources, such as peptone, methionine, tryptophan, sodium nitrate, proline, leucine, tyrosine, valine, aspartic acid, urea, yeast extract, and arginine were individually supplemented at 0.4% into the fermentation medium. Further, the impact of different concentrations (0.1–1.5%) of the optimum nitrogen source was studied to enhance the production of antimicrobial metabolites.27 To evaluate the effect of mineral salts, the optimized medium containing the superior carbon and nitrogen source was supplemented separately with various mineral supplements, such as K2HPO4, KH2PO4, KCl, MgSO4, FeSO4, MnCl2, and NaCl at a concentration of 0.05% (w/v).28
Extraction of the metabolite and antimicrobial activity assayThe strain A. kerguelensis VL-RK_09 was grown under optimized conditions for five days. The culture broth was obtained by centrifugation at 6000rpm for 20min, then extracted with ethyl acetate, and dried in a rotary evaporator at 40°C to obtain a crude extract. The antimicrobial metabolites produced by the strain were tested by an-agar well diffusion assay against bacteria, including S. aureus, B. megaterium, S. flexneri, X. campestris, P. vulgaris, P .aeruginosa, E. coli, Salmonella typhi, Vibrio cholera and Streptococcus mutans (MTCC 497), grown overnight at 37°C, C. albicans (ATCC 10231), Aspergillus niger, A. flavus, Fusarium solani, F. oxysporum (MTCC 3075), Penicillium citrinum and Alternaria sp. were used as test fungi.
Purification of the antimicrobial compoundA seed culture (10%, v/v) prepared by culturing A. kerguelensis VL-RK_09 in ISP-2 (International Sterptomyces Project) medium was transferred to fermentation broth containing malt extract- (1%), lactose (1%), peptone (0.5%), K2HPO4 (0.05%), and CaCO3 (0.2%), with pH adjusted to 7.0. A total of 40 conical flasks of 2-L capacity containing 1L of the broth, were inoculated and incubated on a rotary shaker (180rpm) at 30°C for 120h under optimized conditions. The culture broth (40L) obtained after filtration was extracted twice with ethyl acetate and concentrated under reduced pressure to yield a crude extract (4.2g), which was then subjected to silica gel column (80×2.5cm) chromatography. The separation of the crude extract was conducted by gradient elution with hexane/ethyl acetate. The eluent was run over the column, and small volumes of the eluate collected in test tubes were analyzed by thin-layer chromatography (TLC) using silica gel plates and a hexane/ethyl acetate solvent system. Compounds with identical retention factors (Rf) were combined and assayed for antimicrobial activity against Gram-positive (B. megaterium) and Gram-negative (E. coli) bacteria and the yeast (C. albicans) using an agar plate diffusion assay.23 Among the fractions, the first active fraction (200mg) with good antimicrobial activity was re-chromatographed on a silicagel column using hexane/ethyl acetate (80:20) as an eluting solvent system to obtain 30mg of a pure compound (S1), which was selected for further studies. The structure of the active fraction was deduced by Fourier transform infrared spectroscopy (FT-IR), electrospray ionization mass spectrometry (ESI-MS) and 1H and 13C nuclear magnetic resonance spectroscopy (NMR) spectroscopy.
Biological assaysMinimum inhibitory concentrationThe antimicrobial spectra of the bioactive compound produced by the strain were determined in terms of minimum inhibitory concentration (MIC) by using the agar plate diffusion assay. The MIC of active compound S1 was investigated in comparison with tetracycline. Muller-Hinton agar and Sabouraud dextrose agar media were prepared to grow the bacteria and fungi, respectively. Dilutions of the test compound S1 and the reference drug were prepared in DMSO at concentrations ranging from 0 to 1000μg/mL and were added to cups. The Petri dishes were inoculated with 1.5×104 colony forming units (cfu/mL) and incubated at 37°C for 24–48h for bacteria and 48–72h at 28°C for fungi. The lowest concentration of the bioactive compounds exhibiting significant antimicrobial activity against the test microbes was taken as the MIC of the compound.
The MICs of the bioactive compound (S1) produced by the strain was determined against several pathogenic bacteria and fungi. S. aureus, B. megaterium, B. subtilis (ATCC 6633), B. cereus (MTCC 430), Serratia marcescens (MTCC 1457), X. campestris, P. vulgaris (MTCC 7299), P. aeruginosa (ATCC 9027), E. coli (ATCC 35218), Enterococcus faecalis (MTCC 439), S. mutans, Lactobacillus casei (MTCC 1423), and L. acidophilus (MTCC 495) were used for the antibacterial assay. C. albicans, A. niger, A. flavus, F. solani, F. oxysporum and P. citrinum were used for testing antifungal activity.
Statistical analysisThe results of bioactive metabolite production by A. kerguelensis under different cultural conditions were statistically analyzed with one-way analysis of variance (ANOVA).
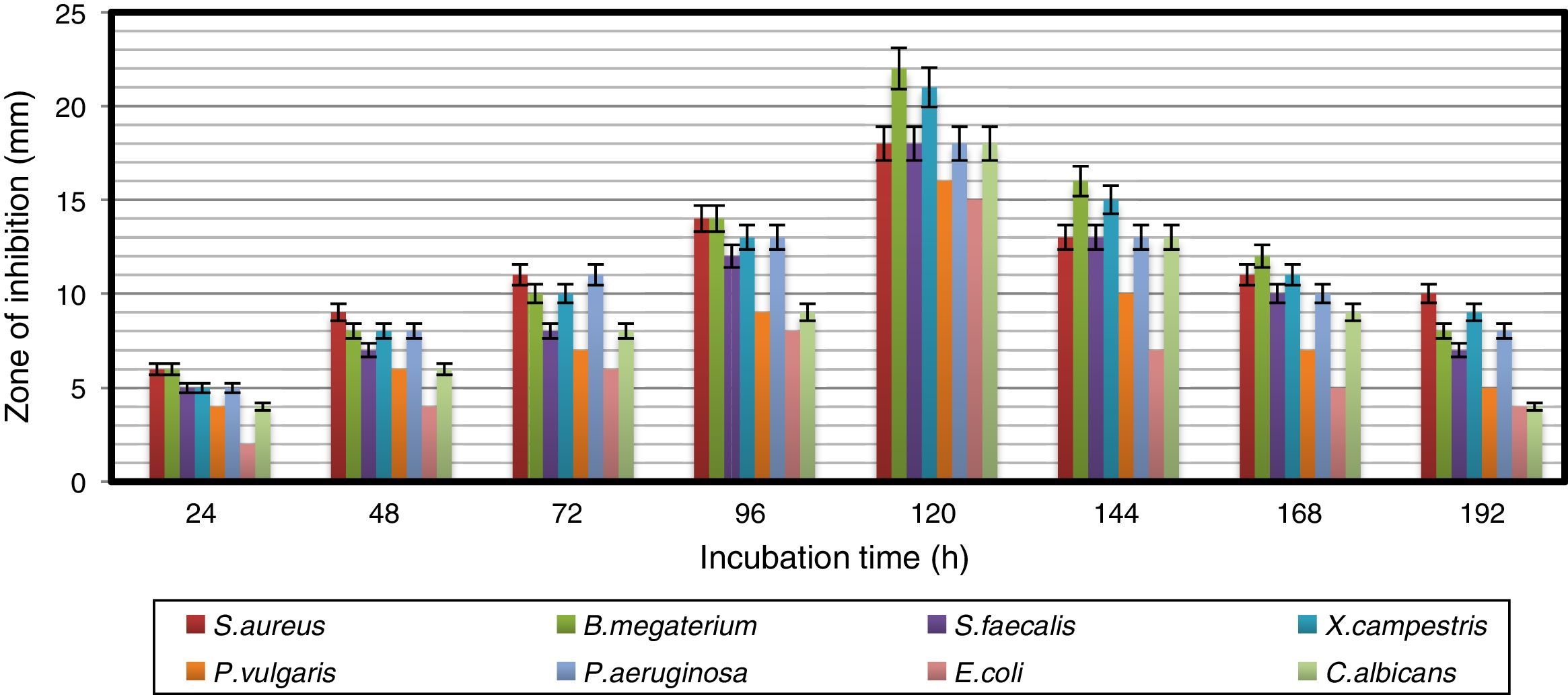
Results and discussionAntimicrobial profile of bioactive metabolites produced by the strainThe growth pattern of A. kerguelensis VL-RK_09 was studied in YMD broth. The stationary phase of the strain lasted from 96 to 120h of incubation (Fig. 1). The crude extract obtained from a five-day-old culture exhibited high antimicrobial activity against the test microorganisms. A previous report demonstrated high antimicrobial activity of crude extracts of a five-day-old culture of Rhodococcus erythropolis VL-RK_05.29 Naragani et al.30 reported the presence of antimicrobial compounds in five-day-old culture extracts of Streptomyces violaceoruber VLK-4.
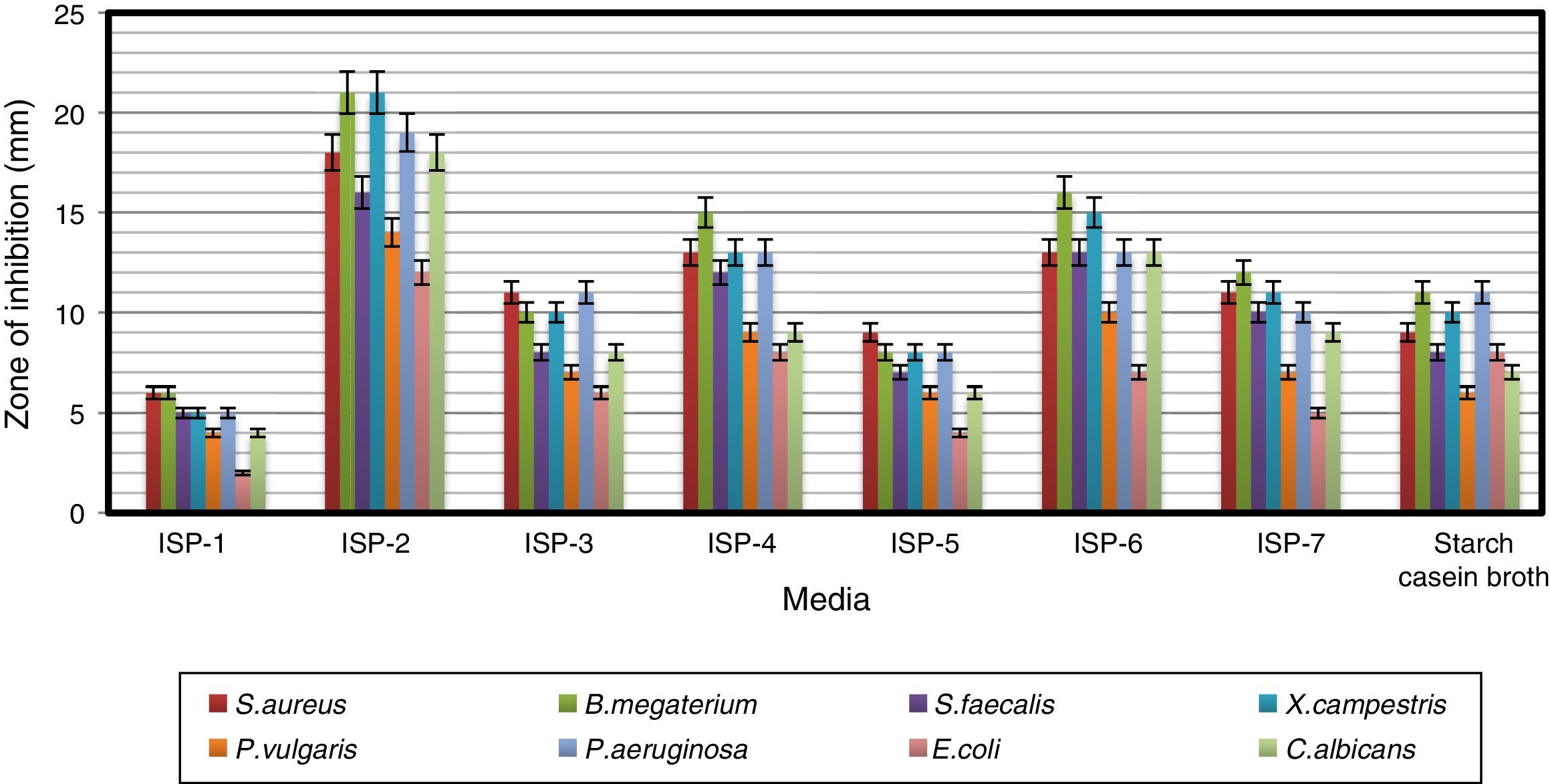
Influence of culture media on antimicrobial activityAntimicrobial activity of the strain was studied by growing it in different culture media (Fig. 2). ISP-2 supported the production of bioactive compounds with the highest antimicrobial activity among the eight media tested, followed by maltose–tryptone broth (ISP-6) and ISP-4. Modified yeast extract–malt extract–dextrose broth (ISP-2) favored the maximum production of bioactive metabolites by Pseudonocardia sp. VUK-10 isolated from mangrove soils.31 Similarly, Naragani et al.30 reported that yeast extract–malt extract–dextrose agar supported the maximum bioactive metabolite production by a marine isolate, R. erythropolis VLK-12.
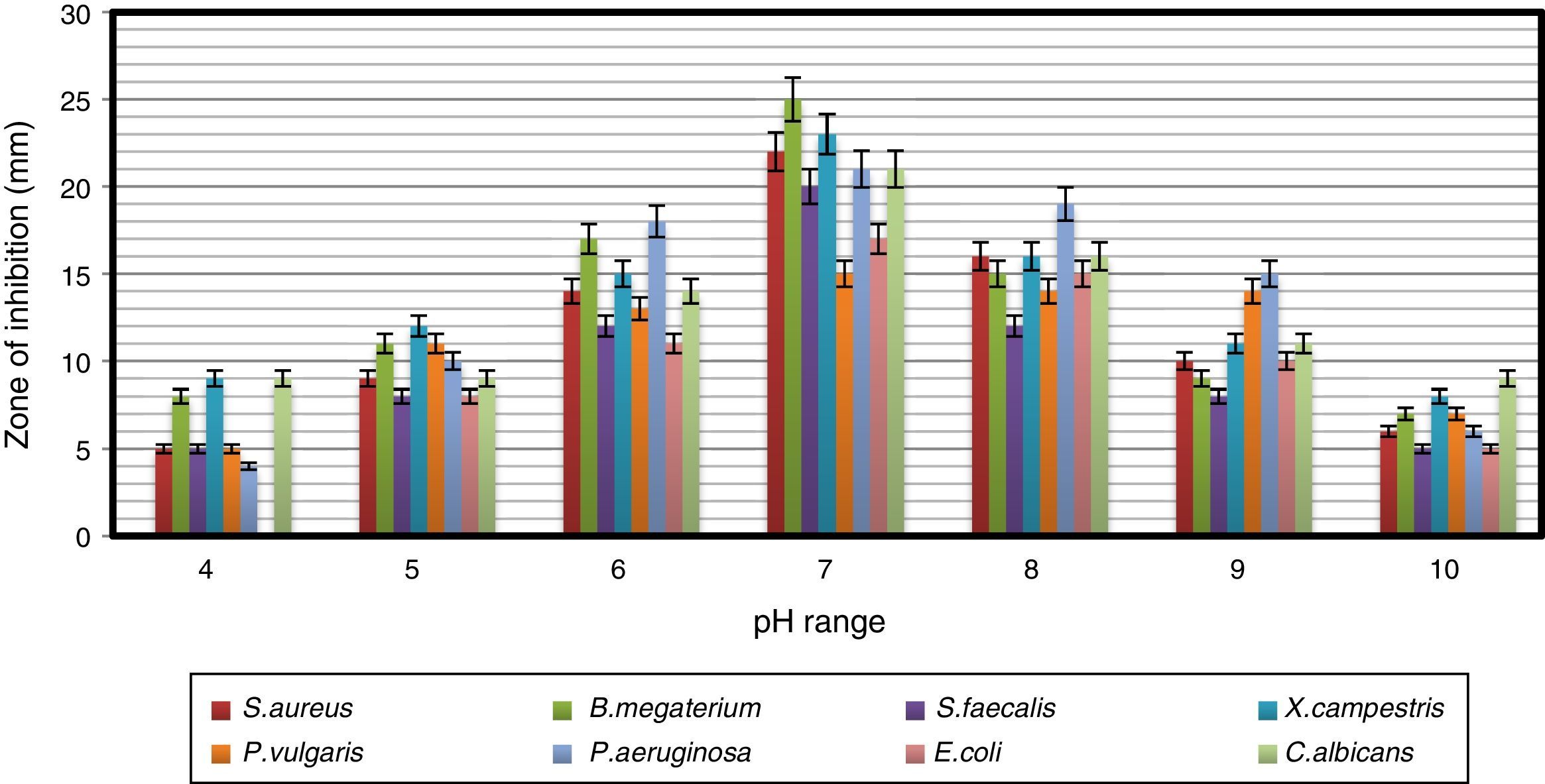
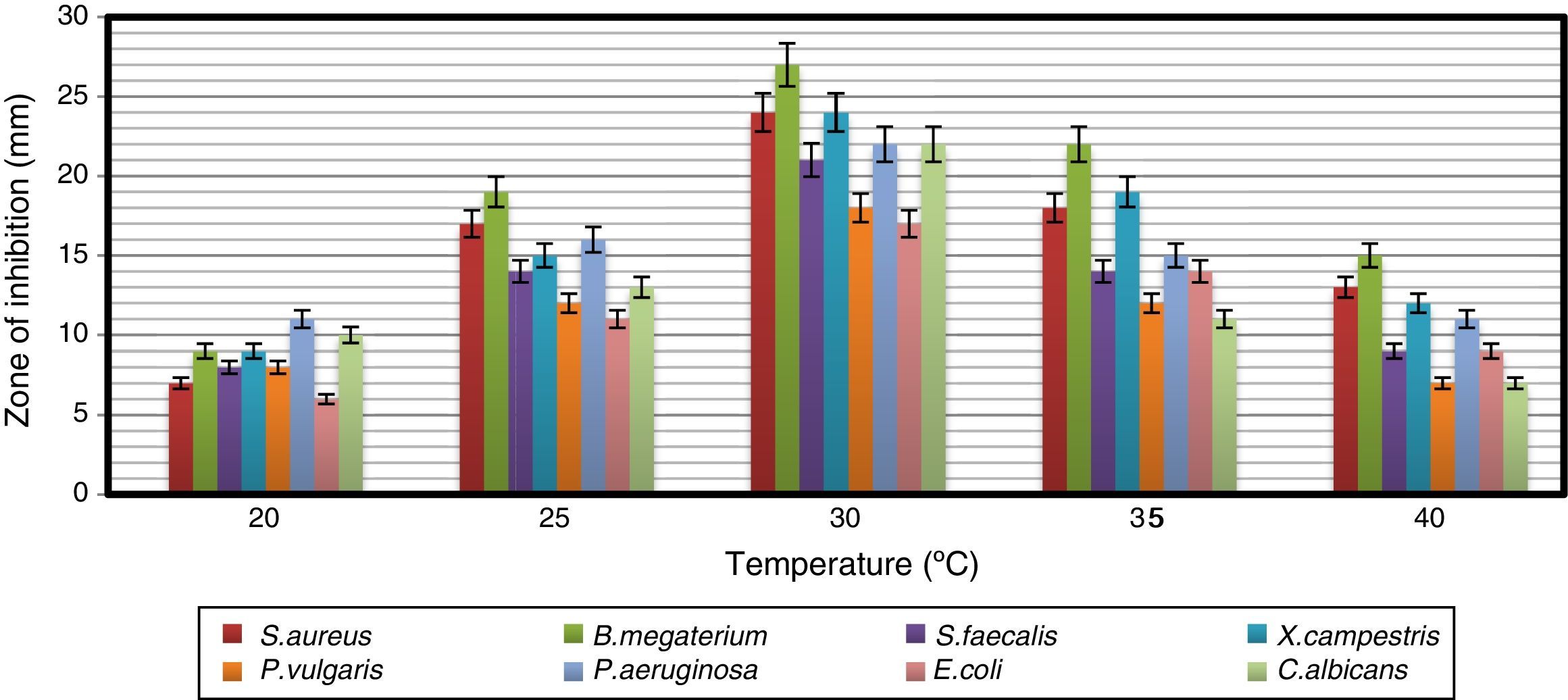
Impact of pH and temperature on antimicrobial activityThe strain was able to grow over a wide range of pH values but showed the maximum antimicrobial activity at pH 7 (Fig. 3). Naragani et al.30 reported high antimicrobial activity of R. erythropolis VLK-12 grown at pH 7. Munaganti et al.29 and Deepa et al.32 reported maximal yields of antimicrobial metabolites produced by R. erythropolis VL-RK_05 and Streptomyces cellulosae VJDS-1 when they were incubated at pH 7. Antimicrobial activity of the strain was also recorded when it was grown at temperatures of 20–40°C, and the optimum was observed at 30°C (Fig. 4). As the incubation temperature increased from 20 to 30°C, there was an increase in bioactive metabolite production. However, a further increase of temperature (above 30°C) led to a decline in the production of bioactive metabolites. Several species of Streptomyces were reported to produce high levels of bioactive metabolites at an incubation temperature of 30°C.33–36
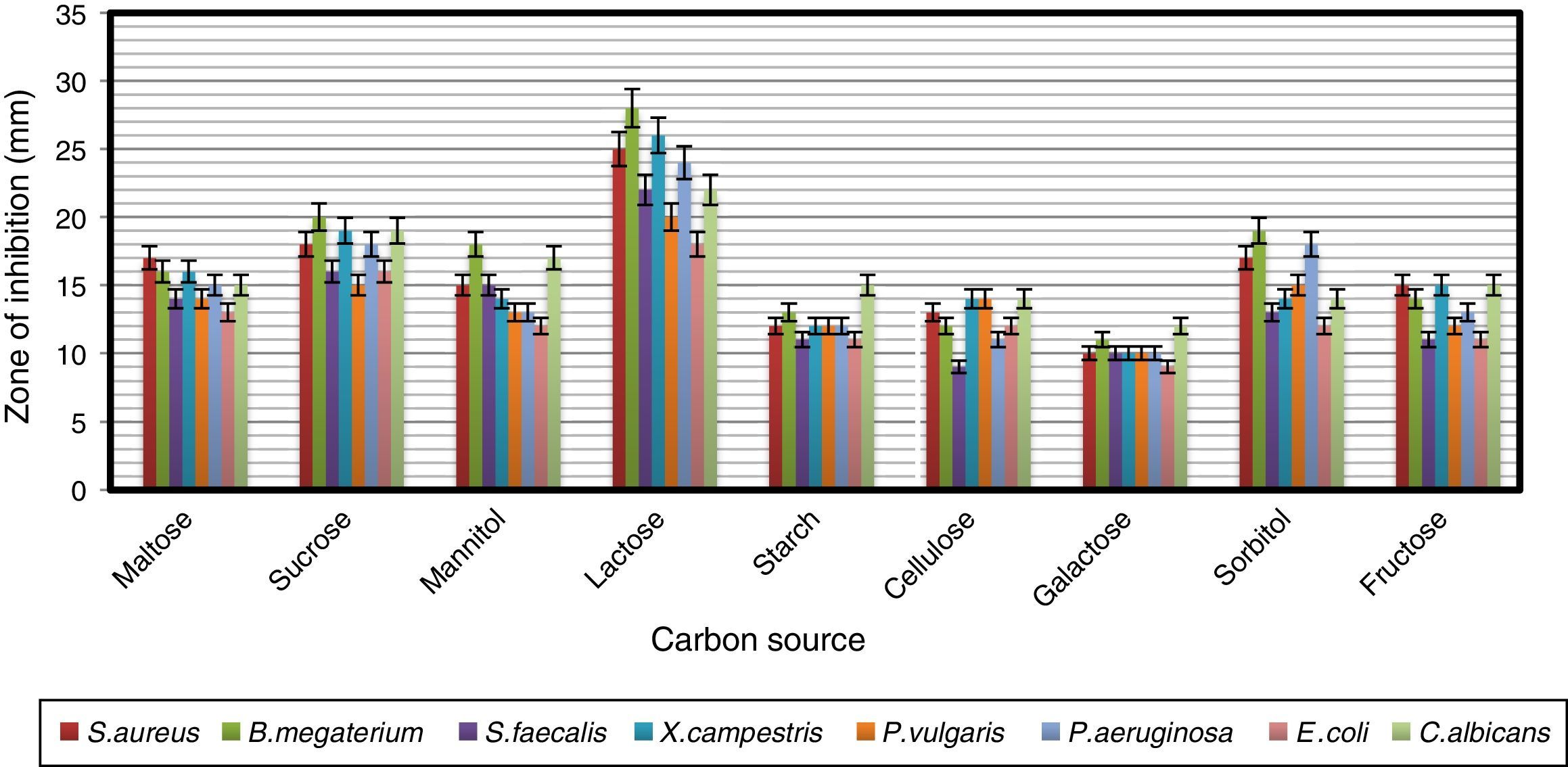
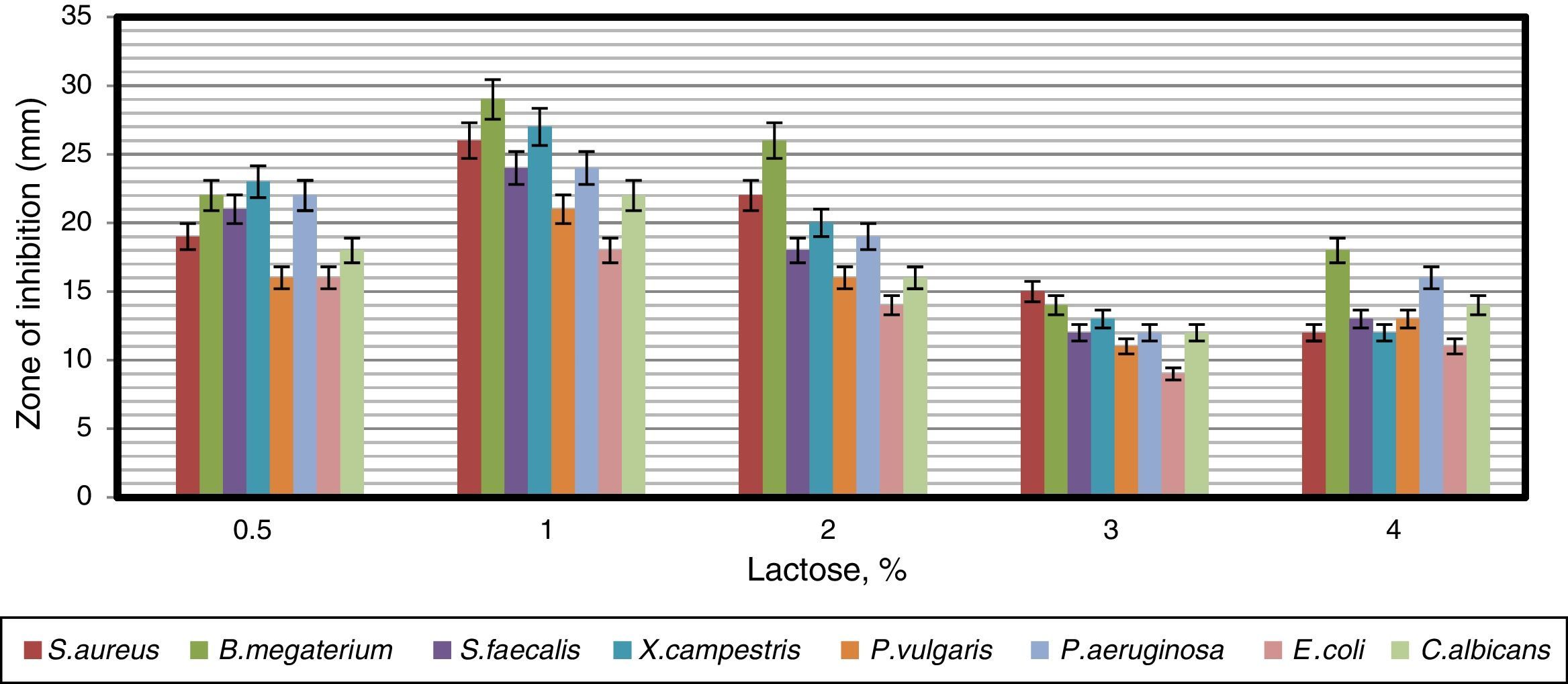
To find a suitable carbon source for secondary metabolite production by the strain, dextrose in YMD broth was replaced with various carbon sources supplemented at a concentration of 0.4%. The effects of carbon sources on production of bioactive metabolites by A. kerguelensis VL-RK_09 are presented in Fig. 5. Since lactose supported a high yield of bioactive metabolites, different concentrations of lactose (0.5–4%) were tested to determine the optimal concentration. Lactose at a concentration of 1% supported the highest yield of bioactive metabolites (Fig. 6).
These results are comparable with those obtained for Streptomyces hygroscopicus strains AK-111-81 and CH-7, which utilized lactose as a carbon source for antibiotic production.37,38
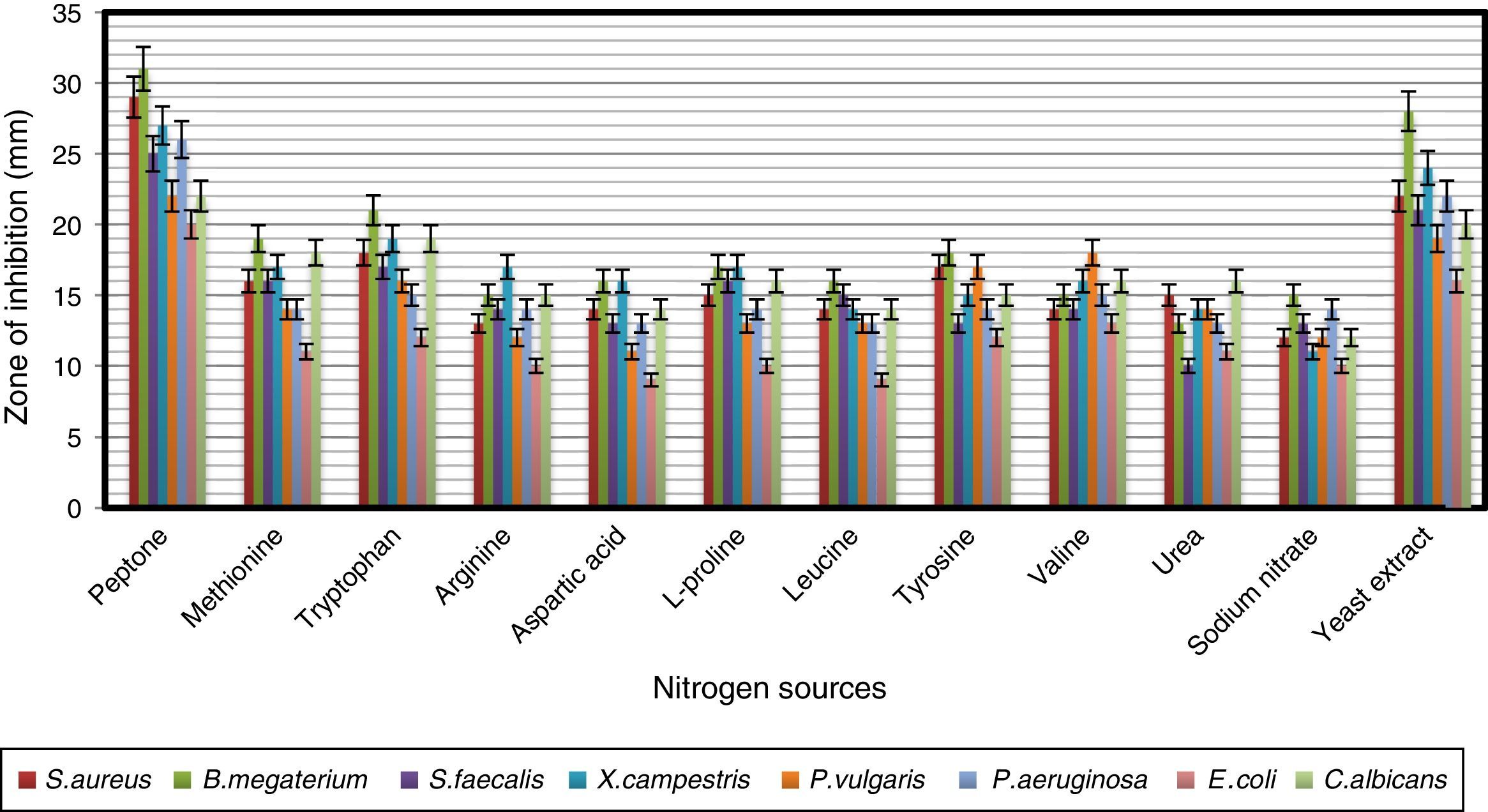
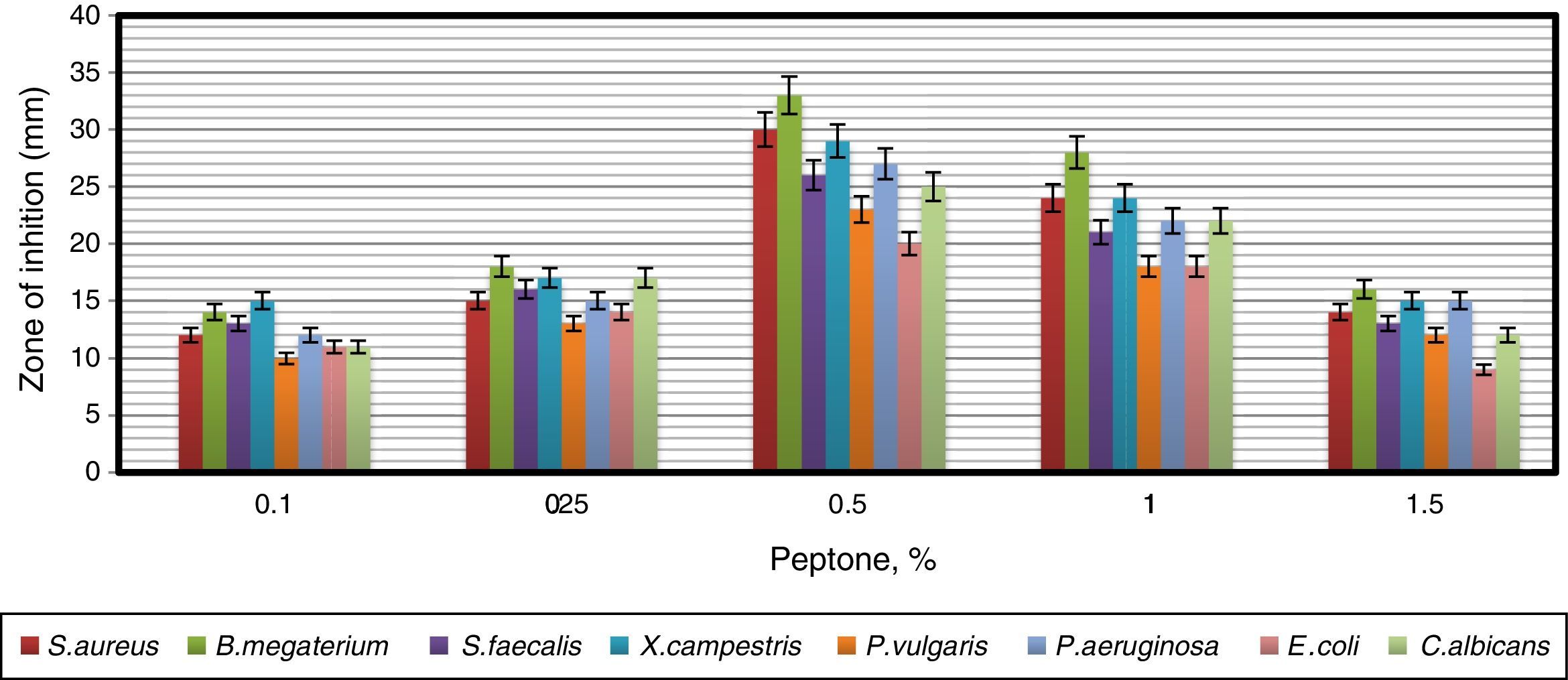
In order to obtain an effective composition of the growth medium, various nitrogen sources were evaluated for their influence on antimicrobial activity of the strain. Among the nitrogen sources tested, peptone was found to be the best for antimicrobial metabolite, followed by yeast extract and tryptophan (Fig. 7). Since peptone enhanced the antimicrobial activity of strain VL-RK_09, the effects of different concentrations of peptone were tested, and 0.5% was found to be the best for improved antimicrobial activity (Fig. 8). Peptone was also reported as a suitable nitrogen source for obtaining optimal yields of bioactive metabolites produced by Streptomyces rochei G16439 and S. scabies PK-A41.40
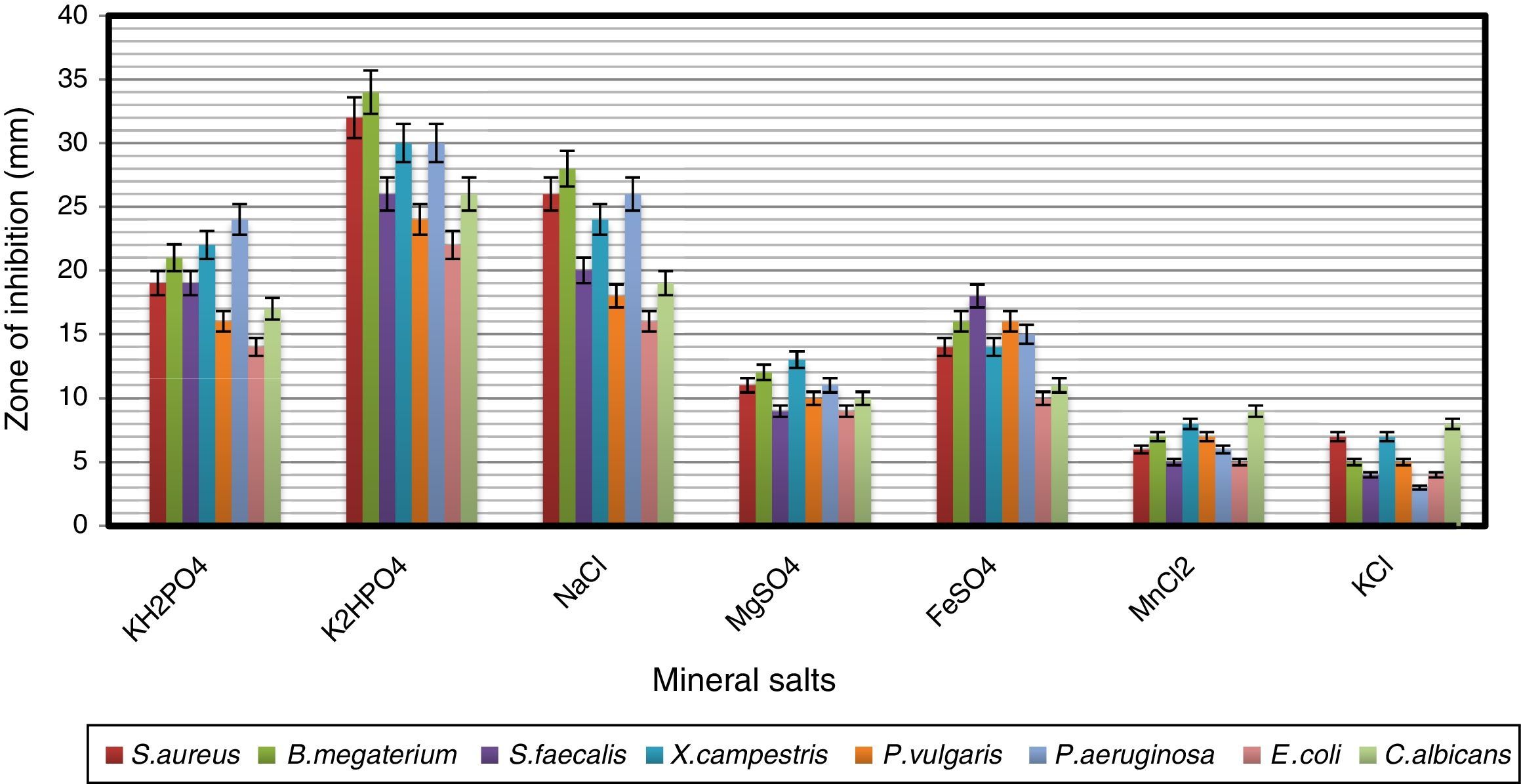
The effects of mineral salts on secondary metabolite production by strain VL-RK_09 are shown in Fig. 9. Among the mineral salts tested, K2HPO4 supported the highest antimicrobial activity. Similar results were reported for Streptomyces albidoflavus.41 Ripa et al.31 and Usha et al.42 reported that K2HPO4 showed positive effects on antibiotic production by Streptomyces sp. RUPA-08PR and Pseudonocardia sp.
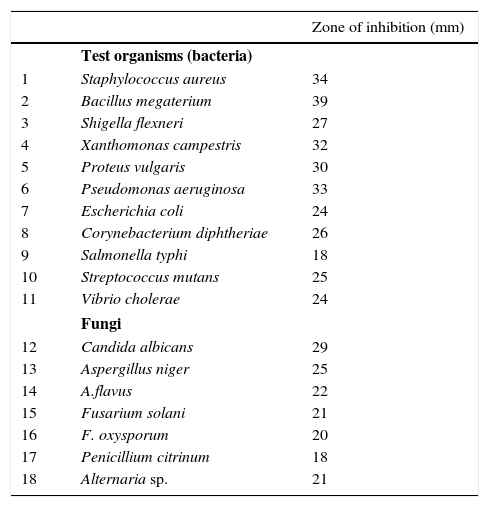
The secondary metabolites obtained from the strain VL-RK_09 under optimized cultural conditions, which included malt extract (1%), lactose (1%), peptone (0.5%), K2HPO4 (0.05%), CaCO3 (0.2%), pH 7.0±0.2, temperature 30°C, and the incubation period of five days, showed strong antimicrobial activity against bacteria and fungi (Table 1). Among the bacteria tested, B. megaterium, S. aureus, P. aeruginosa, X. campestris, and P. vulgaris appeared to be highly sensitive to the metabolites produced by the strain.
Antimicrobial activity of Arthrobacter kerguelensis VL-RK_09 under optimized culturing conditions.
| Zone of inhibition (mm) | ||
|---|---|---|
| Test organisms (bacteria) | ||
| 1 | Staphylococcus aureus | 34 |
| 2 | Bacillus megaterium | 39 |
| 3 | Shigella flexneri | 27 |
| 4 | Xanthomonas campestris | 32 |
| 5 | Proteus vulgaris | 30 |
| 6 | Pseudomonas aeruginosa | 33 |
| 7 | Escherichia coli | 24 |
| 8 | Corynebacterium diphtheriae | 26 |
| 9 | Salmonella typhi | 18 |
| 10 | Streptococcus mutans | 25 |
| 11 | Vibrio cholerae | 24 |
| Fungi | ||
| 12 | Candida albicans | 29 |
| 13 | Aspergillus niger | 25 |
| 14 | A.flavus | 22 |
| 15 | Fusarium solani | 21 |
| 16 | F. oxysporum | 20 |
| 17 | Penicillium citrinum | 18 |
| 18 | Alternaria sp. | 21 |
The culture filtrate obtained after 120h of fermentation was extracted with ethyl acetate and concentrated in vacuum to yield a dark brown residue, which was then subjected to silica gel column chromatography using a gradient solvent system of hexane/ethyl acetate.
Among the fractions, the first active fraction (200mg) with good antimicrobial activity was re-chromatographed on a silica gel column using hexane: ethyl acetate (80:20) as the eluting solvent system to obtain a pure compound (30mg). The structure of the purified compound S1 was elucidated and characterized by FT-IR, mass, and NMR spectroscopy.
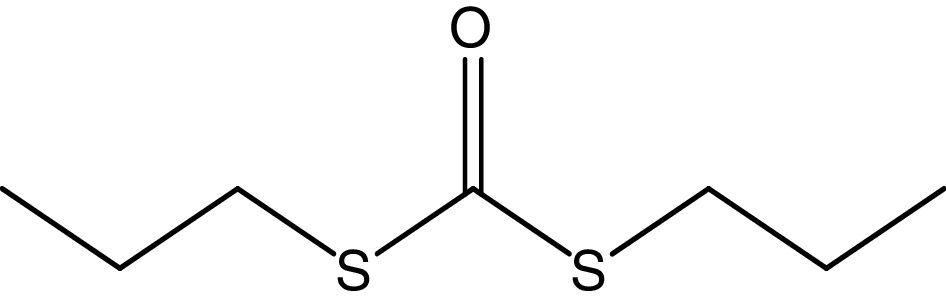
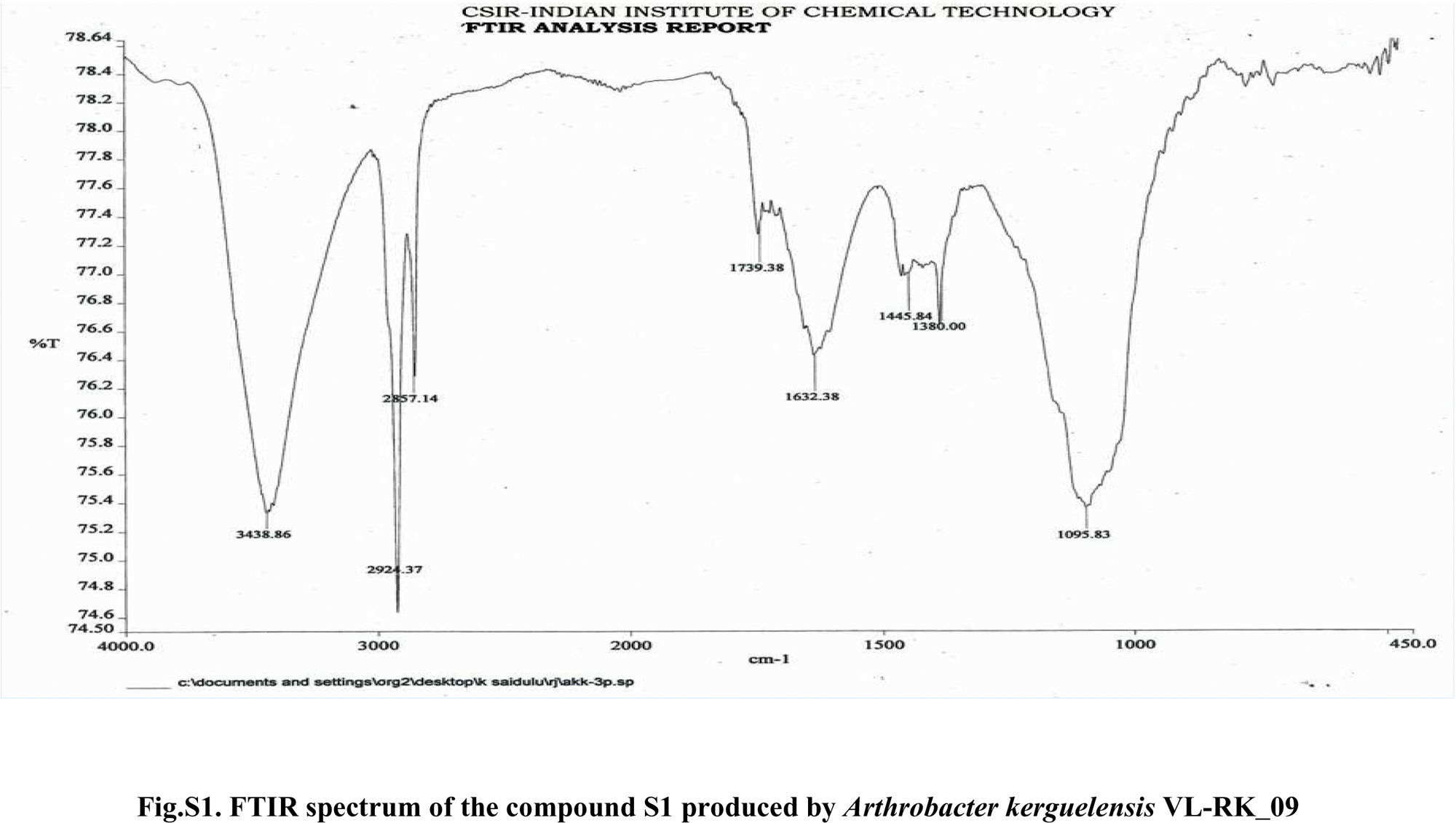
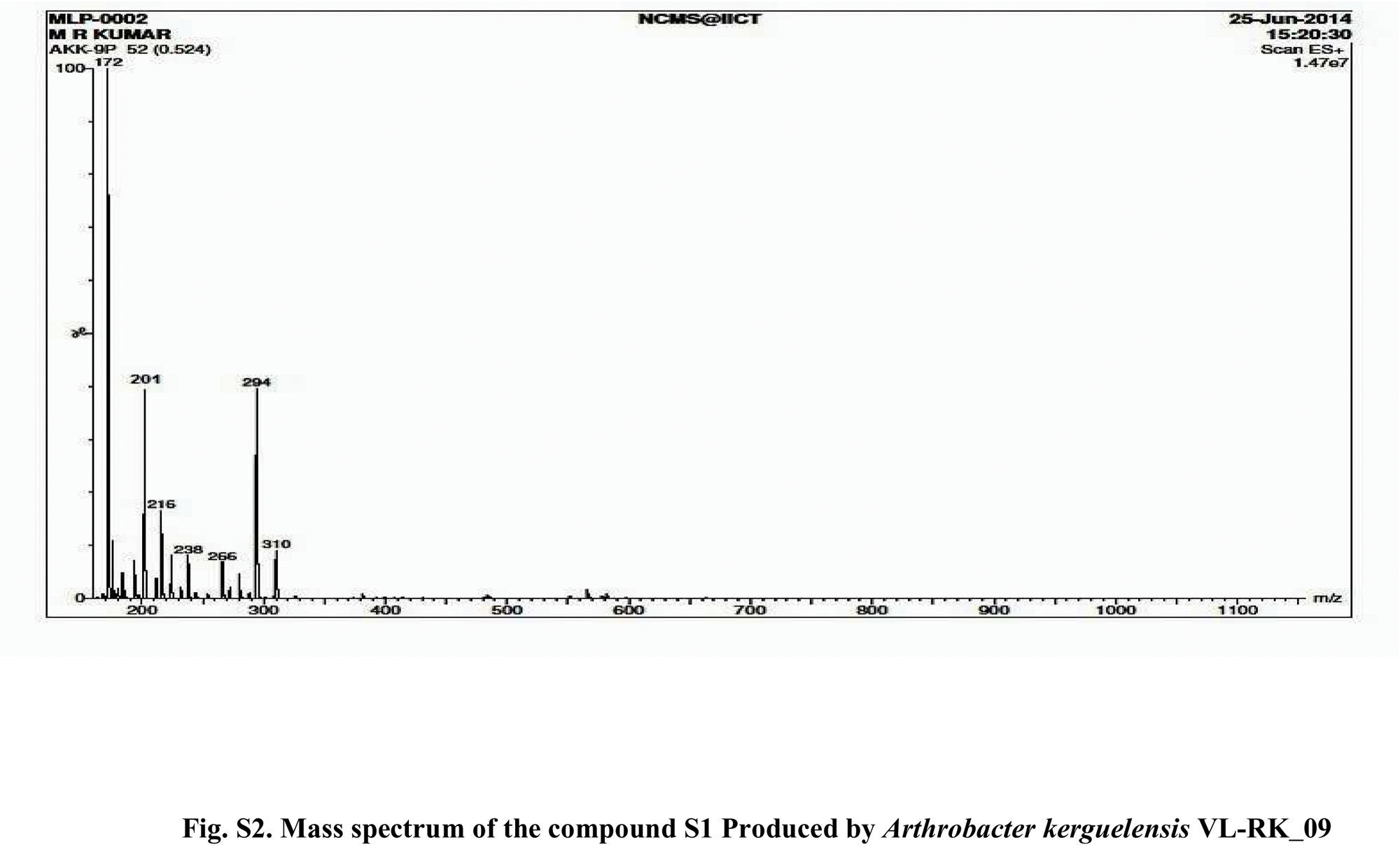
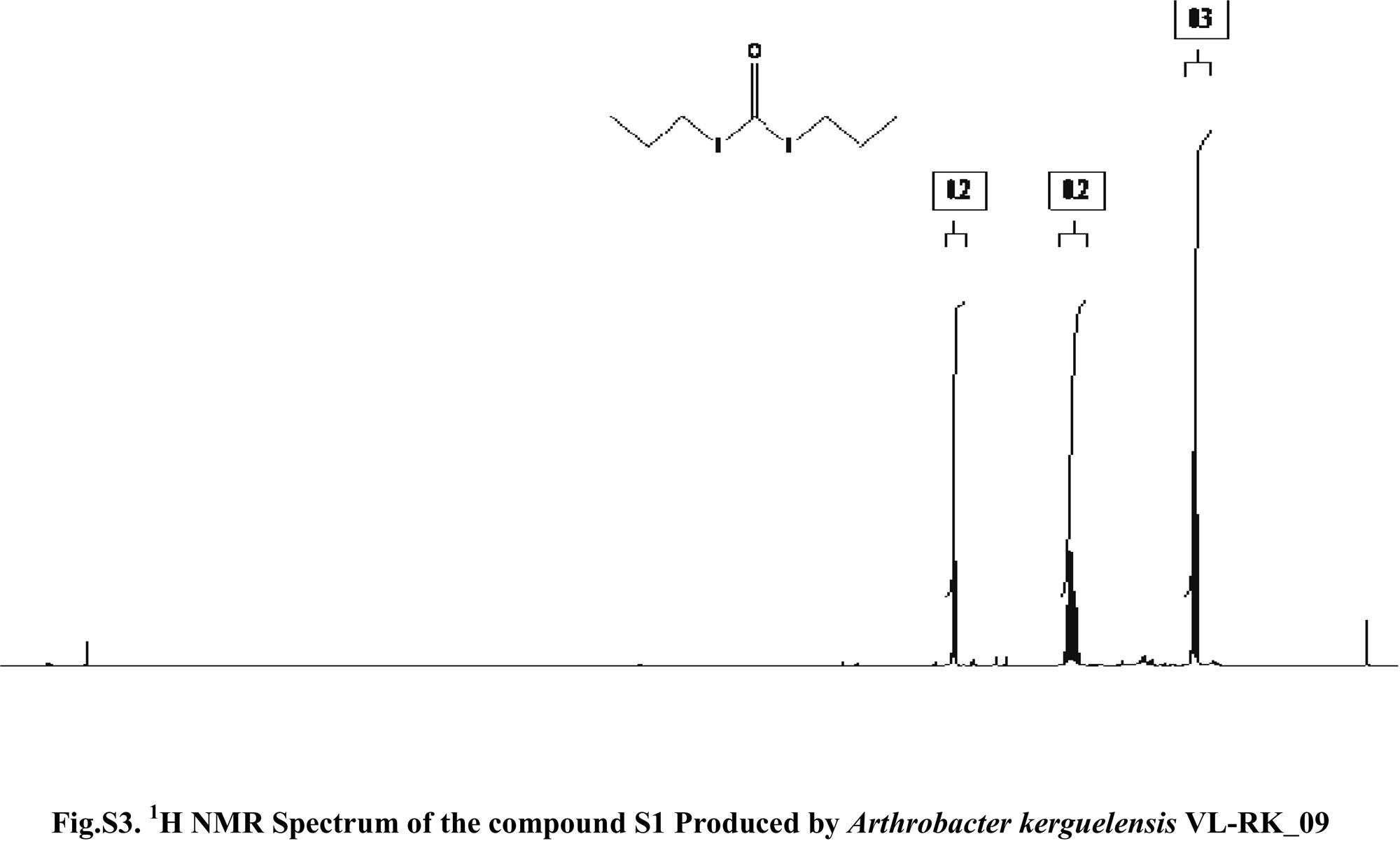
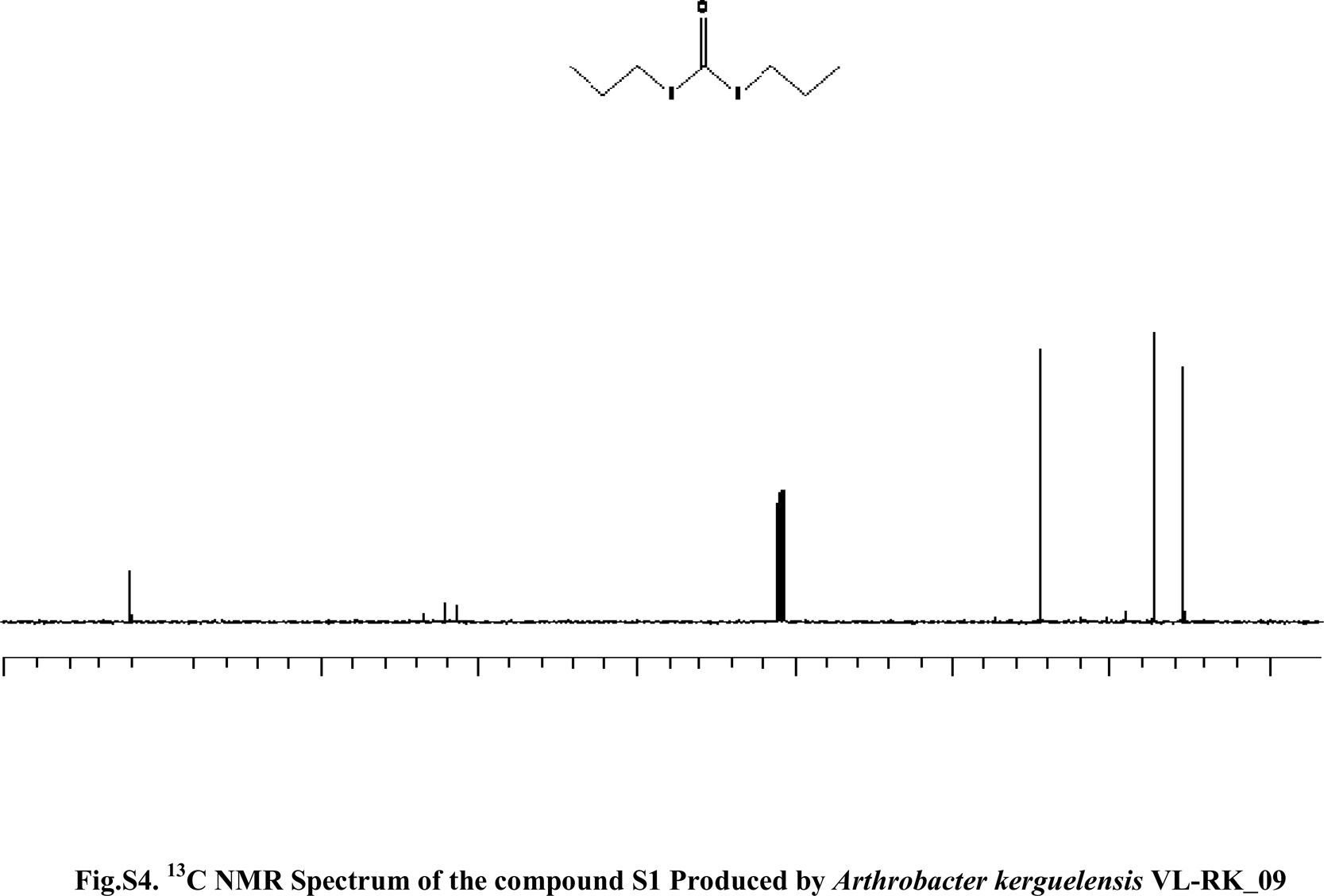
Compound S1 was isolated as a yellow-orange liquid, soluble in CHCl3, methanol, dichloromethane, and DMSO. The IR absorption maximums (Vmax) at 2924, 2856, 1739, 1629, 1380 and 1095cm−1 suggested the presence of carbonyl functional groups (Fig. S1). In ESI–MS, the compound showed molecular ions at m/z 178, inferring the molecular formula of C7H14OS2 (Fig. S2). The proton NMR of the compound displayed proton signals at 2.33 (t, J=7.3Hz, 2H), 1.66 (q, J=7.3, 7.47Hz, 2H), and 0.97 (t, J=7.47Hz, 3H) (Fig. S3). 13C NMR depicted peaks and showed signals at 180.12, 35.97, 17.73, and 13.34 (Fig. S4). Based on these spectral data, the first active compound S1 was identified as S,S-dipropyl carbonodithioate (Fig. 10). This is the first report of the occurrence of this compound in the genus Arthrobacter.
Arthrobacter is a common genus of soil bacteria, which are metabolically versatile and produce different enzymes allowing the bacteria to grow on a wide range of substrates. The members of the genus are not well explored as secondary metabolite producers although there had been reports of production of bioactive compounds by some species. Antimicrobial metabolites have been studied in some aquatic and polar strains of Arthrobacter.43,44 Hence, an attempt was made in the present study to evaluate the production of antimicrobial metabolites by a terrestrial isolate, A. kerguelensis VL-RK_09, isolated from a mango orchard.
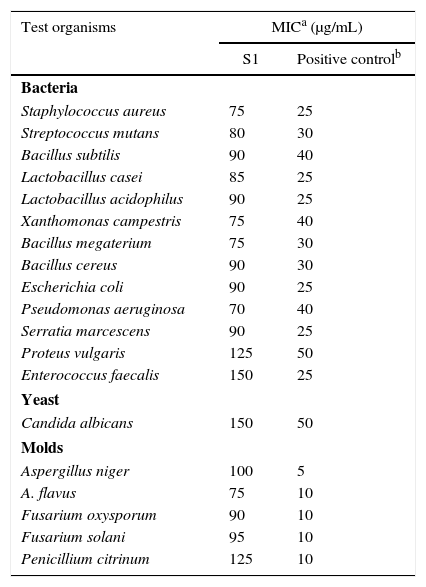
Biological assayThe antimicrobial profile of the bioactive compound based on the MIC values is shown in Table 2. B. megaterium was highly sensitive to the compound, followed by S. aureus, S. mutans, B. subtilis, and B. cereus among the Gram-positive bacteria. P. aeruginosa was highly sensitive to the compound followed by X. campestris, E. coli, S. marcesens, P. vulgaris, and E. faecalis among the Gram-negative bacteria.
MIC of S,S-dipropyl carbonodithioate isolated from Arthrobacter kerguelensis VL-RK_09 against different pathogenic bacteria and fungi.
| Test organisms | MICa (μg/mL) | |
|---|---|---|
| S1 | Positive controlb | |
| Bacteria | ||
| Staphylococcus aureus | 75 | 25 |
| Streptococcus mutans | 80 | 30 |
| Bacillus subtilis | 90 | 40 |
| Lactobacillus casei | 85 | 25 |
| Lactobacillus acidophilus | 90 | 25 |
| Xanthomonas campestris | 75 | 40 |
| Bacillus megaterium | 75 | 30 |
| Bacillus cereus | 90 | 30 |
| Escherichia coli | 90 | 25 |
| Pseudomonas aeruginosa | 70 | 40 |
| Serratia marcescens | 90 | 25 |
| Proteus vulgaris | 125 | 50 |
| Enterococcus faecalis | 150 | 25 |
| Yeast | ||
| Candida albicans | 150 | 50 |
| Molds | ||
| Aspergillus niger | 100 | 5 |
| A. flavus | 75 | 10 |
| Fusarium oxysporum | 90 | 10 |
| Fusarium solani | 95 | 10 |
| Penicillium citrinum | 125 | 10 |
Among the fungi tested, A. flavus was found to be more sensitive than the others. The MIC values of S,S-dipropyl carbonodithioate against the test bacteria and fungi varied from 75 to 150μg/mL.
ConclusionsIn the present study, a rare actinobacterium was isolated on ISP-2 agar medium as a predominant strain from soil collected at a mango orchard. Screening of the strain for antagonistic activity revealed its potential to inhibit several opportunistic pathogens in vitro. The strain was identified as A. kerguelensis by genomic analysis and designated as VL-RK_09. In order to improve the productivity of the strain, an optimization of nutritional and physiological factors was carried out. The strain exhibited high antimicrobial activity when grown in an optimized culture medium (lactose, 1%; peptone, 0.5%; K2HPO4, 0.05%) at pH 7.0 and incubated at 30°C for 120h. An attempt was also made to characterize antimicrobial metabolites by culturing the strain in a large volume of the medium. Extraction, followed by bioactivity-guided isolation and purification of a crude ethylacetate extract, revealed the presence of S,S-dipropyl carbonodithioate. This is the first report on S,S-dipropyl carbonodithioate isolated from A. kerguelensis VL-RK_09.
Conflicts of interestThe authors declare no conflicts of interest.
We would like to thank the Department of Botany and Microbiology for providing the laboratory facilities. RKM thanks UGC for providing the financial assistance to carry out the research work.