Staphylococcus spp. play an important role in the etiology of bovine mastitis. Staphylococcus aureus is considered the most relevant species due to the production of virulence factors such as slime, which is required for biofilm formation. This study aimed to evaluate biofilm production and its possible relation to beta-lactamic resistance in 20 S. aureus isolates from bovine mastitic milk. The isolates were characterized by pheno-genotypic and MALDI TOF-MS assays and tested for genes such as icaA, icaD, bap, agr RNAIII, agr I, agr II, agr III, and agr IV, which are related to slime production and its regulation. Biofilm production in microplates was evaluated considering the intervals determined along the bacterial growth curve. In addition, to determine the most suitable time interval for biofilm analysis, scanning electron microscopy was performed. Furthermore, genes such as mecA and blaZ that are related to beta-lactamic resistance and oxacillin susceptibility were tested. All the studied isolates were biofilm producers and mostly presented icaA and icaD. The Agr type II genes were significantly prevalent. According to the SEM, gradual changes in the bacterial arrangement were observed during biofilm formation along the growth curve phases, and the peak was reached at the stationary phase. In this study, the penicillin resistance was related to the production of beta-lactamase, and the high minimal bactericidal concentration for cefoxitin was possibly associated with biofilm protection. Therefore, further studies are warranted to better understand biofilm formation, possibly contributing to our knowledge about bacterial resistance in vivo.
Staphylococcus spp. play an important role in the etiology of intramammary infections of dairy cattle. Staphylococcus aureus stands out among the prevalent etiologic agents in this type of infection due to its ability to produce a wide array of virulence factors that contribute to the bacterial invasion.1 Of them, the production of slime, an extracellular mucopolysaccharide, appears to play a crucial role in the adhesion and colonization of the microorganism on the mammary glandular epithelium; this not only favors biofilm formation and their extracellular persistence but also ensures success in their installation and maintenance in the host tissues.2 Slime is composed of a high-molecular-weight polysaccharide intercellular adhesin. Its production is mediated by the intercellular adhesion operon (ica) formed by the genes icaA, icaB, icaC, and icaD and a regulator gene, icaR, which encodes the ICAA, ICAB, ICAC, and ICAD proteins.3
Furthermore, ica-independent mechanisms possibly play an essential role in bacterial biofilm formation. For example, the function of bap, which encodes for the surface protein Bap, is to assist in intercellular adhesion and biofilm formation. This gene has been primarily studied in isolates from bovine mastitis.4
The repression of agr quorum-sensing system is necessary for biofilm formation. Its reactivation in established biofilms through auto inducing peptides (AIPs) addition or glucose depletion triggers biofilm detachment.5 The agr system includes AgrD, the signaling octapeptide produced in high cell density; AgrB, a transmembrane protein responsible for secretion, export, and processing of active AgrD; and AgrC, a membrane receptor that triggers AgrA phosphorylation mechanism when bound to AgrD. The phosphorylated AgrA positively regulates the production of the effector molecule RNA III.6S. aureus can be classified into four polymorphic Agr types (AgrI, AgrII, AgrIII, and AgrIV) based on the specificity of AIP to the signal receptor AgrC.7
Moreover, biofilm production in S. aureus from mastitis can be associated with antimicrobial resistance.8 The mechanisms responsible for this resistance include the physical and chemical diffusion barrier formed by the exopolysaccharide matrix, which hinders antimicrobial penetration, the existence of microenvironments that antagonize the antibiotic action, the activation of stress responses that cause changes in bacterial physiology, and the stable and slower growth of these microorganisms due to nutrient limitation and the absence of antimicrobial targets.2
Antimicrobials such as beta-lactams are preferred for the treatment of staphylococcal infections. However, production of beta-lactamase enzymes, coded by blaZ that hydrolyzes the beta-lactamic ring, and production of low-affinity penicillin binding protein (PBP2a), coded by mecA, may lead to antimicrobial resistance.9
This study aimed to detect the phenotypic expression of biofilm and the presence of structural and regulatory genes involved in the production of this virulence factor. In addition, the stages of biofilm synthesis along the growth curve were evaluated by scanning electron microscopy (SEM), and pheno-genotypic resistance to beta-lactamic and its possible relation to biofilm production were evaluated.
Materials and methodsSampling and pheno-genotypic and proteomic identificationThree dairy cattle farms located in an important milk production region of Rio de Janeiro, Brazil, were selected owing to the high prevalence of subclinical mastitis on the farms, identified through the California mastitis test and somatic cell count. In total, 120 milk samples were collected in October and November 2012. Fifty nine Staphylococcus spp. were isolated, of which 41 were S. aureus strains.
After phenotypic identification, all 41 strains were submitted to polymerase chain reaction (PCR) for 16S rRNA to confirm the Staphylococcus spp.10 PCR for coa,11nuc,12 and 23S rDNA13 genes were performed to characterize S. aureus. The ATCC 29213 S. aureus was used as quality control. Furthermore, all isolates were evaluated by the matrix-assisted laser desorption ionization-time of flight mass spectrometry, as described by Motta et al.,14 considering the accepted values for matches ≥2.
The S. aureus isolates were subjected to disk diffusion tests using amoxicillin (10μg), ampicillin (10μg), azithromycin (15μg), ciprofloxacin (5μg), chloramphenicol (30μg), cefepime (30μg), enrofloxacin (5mcg), erythromycin (15μg), streptomycin (10μg), moxifloxacin (5μg), neomycin (30mcg), novobiocin (5mcg), cotrimoxazole (25μg), and tetracycline (30μg) disks. After overnight incubation at 35°C, followed by inhibition zone measurement, the results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) standards.15–17 These isolates were subjected to DNA extraction and amplification of hlA and hlB,18fbnA and fbnB,19 and cap5 and cap8,19 according to the protocol described by Marques et al.20 and Tito et al.21 To study the biofilm production, 20 S. aureus strains were selected considering their antibiotic resistance profiles and the presence of virulence genes.
Qualitative and quantitative biofilm assayBiofilm production was measured using qualitative and quantitative assays, described by Marques et al.20 All the 20 S. aureus isolates were transferred to sheep blood agar for 24h at 35°C. The grown colonies were inoculated into tryptic soy broth (TSB) containing 0.24% glucose to stimulate slime production for 24h at 35°C. The bacterial cultures were adjusted to a 0.5 McFarland scale and diluted 1:10 in TSB with the addition of 0.24% glucose. Aliquots of this suspension (200μL) were inoculated into sterile polystyrene 96-wellmicroplates for 24h at 35°C without agitation. After discarding this material, the wells were washed twice with 200μL sterile saline, oven dried at 65°C for 1h, and stained with 200μL safranin 1% for 15min. Subsequently, the wells were washed three times with distilled water and dried at room temperature. The absorbance was determined at 490nm in an ELISA reader (BIO RAD MODEL 680). Uninoculated wells containing TSB broth with 0.24% glucose were used as controls. The tests were performed in triplicate. The strains were classified according to the following OD values: strong ≥0.3, moderate ≥0.2 and <0.3, weak ≥0.1 and <0.2, and negative <0.1.20
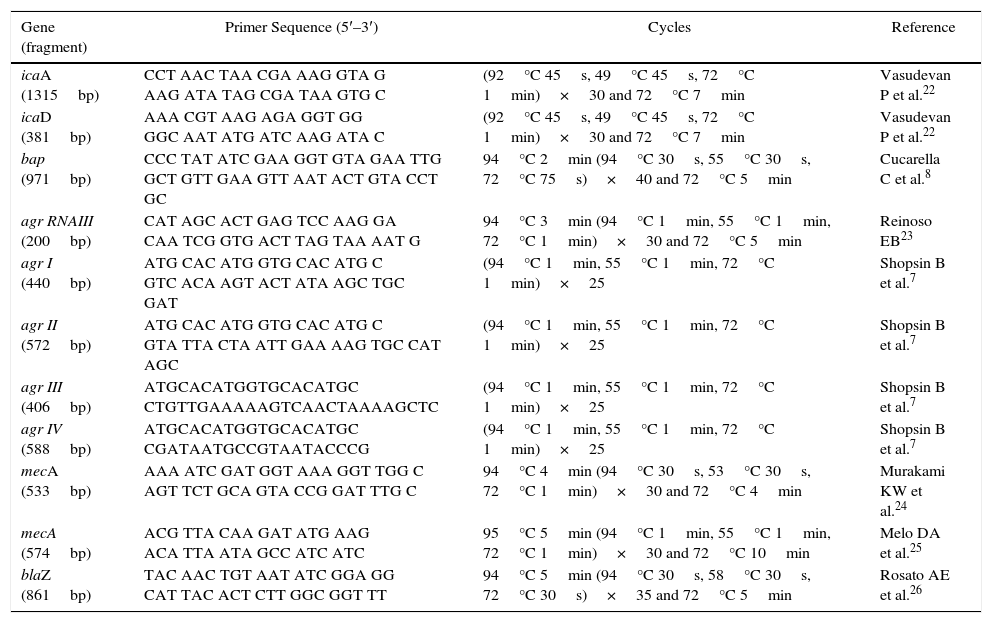
Biofilm gene and Agr typesAll isolates were subjected to DNA extraction and amplification of icaA and icaD,22bap,8 and agr RNAIII,23 according to the protocol described by Marques et al.20 and Tito et al.21 (Table 1).
Primers and amplification conditions for the detection of resistance, biofilm genes, and agr genes.
| Gene (fragment) | Primer Sequence (5′–3′) | Cycles | Reference |
|---|---|---|---|
| icaA (1315bp) | CCT AAC TAA CGA AAG GTA G AAG ATA TAG CGA TAA GTG C | (92°C 45s, 49°C 45s, 72°C 1min)×30 and 72°C 7min | Vasudevan P et al.22 |
| icaD (381bp) | AAA CGT AAG AGA GGT GG GGC AAT ATG ATC AAG ATA C | (92°C 45s, 49°C 45s, 72°C 1min)×30 and 72°C 7min | Vasudevan P et al.22 |
| bap (971bp) | CCC TAT ATC GAA GGT GTA GAA TTG GCT GTT GAA GTT AAT ACT GTA CCT GC | 94°C 2min (94°C 30s, 55°C 30s, 72°C 75s)×40 and 72°C 5min | Cucarella C et al.8 |
| agr RNAIII (200bp) | CAT AGC ACT GAG TCC AAG GA CAA TCG GTG ACT TAG TAA AAT G | 94°C 3min (94°C 1min, 55°C 1min, 72°C 1min)×30 and 72°C 5min | Reinoso EB23 |
| agr I (440bp) | ATG CAC ATG GTG CAC ATG C GTC ACA AGT ACT ATA AGC TGC GAT | (94°C 1min, 55°C 1min, 72°C 1min)×25 | Shopsin B et al.7 |
| agr II (572bp) | ATG CAC ATG GTG CAC ATG C GTA TTA CTA ATT GAA AAG TGC CAT AGC | (94°C 1min, 55°C 1min, 72°C 1min)×25 | Shopsin B et al.7 |
| agr III (406bp) | ATGCACATGGTGCACATGC CTGTTGAAAAAGTCAACTAAAAGCTC | (94°C 1min, 55°C 1min, 72°C 1min)×25 | Shopsin B et al.7 |
| agr IV (588bp) | ATGCACATGGTGCACATGC CGATAATGCCGTAATACCCG | (94°C 1min, 55°C 1min, 72°C 1min)×25 | Shopsin B et al.7 |
| mecA (533bp) | AAA ATC GAT GGT AAA GGT TGG C AGT TCT GCA GTA CCG GAT TTG C | 94°C 4min (94°C 30s, 53°C 30s, 72°C 1min)×30 and 72°C 4min | Murakami KW et al.24 |
| mecA (574bp) | ACG TTA CAA GAT ATG AAG ACA TTA ATA GCC ATC ATC | 95°C 5min (94°C 1min, 55°C 1min, 72°C 1min)×30 and 72°C 10min | Melo DA et al.25 |
| blaZ (861bp) | TAC AAC TGT AAT ATC GGA GG CAT TAC ACT CTT GGC GGT TT | 94°C 5min (94°C 30s, 58°C 30s, 72°C 30s)×35 and 72°C 5min | Rosato AE et al.26 |
The agr system groups were classified based on the hypervariable domain of the agr locus, according to Shopsin et al.7 A forward primer, pan-agr, corresponding to the conserved sequences of agrB, was used in all the reactions. Furthermore, four reverse primers were used, each specific for the amplification of a single group of agr, based on the agr locus polymorphism. Duplex PCR was performed to classify the groups based on the following products: Agr I (440bp) and Agr II (572bp) and Agr III (406bp) and Agr IV (588bp) (Table 1). PCR products were separated by electrophoresis on 1% agarose gels using SYBR Green (Invitrogen®) diluted dye (1:100). The amplicons were visualized and documented using the image capturing system L-PIX EX (Loccus Biotecnologia®).
Growth curve estimationA 1-mL aliquot of bacterial culture [106CFU (colony forming unit)/mL] was diluted ten-fold in a simple broth (0.4% meat extract; 1% peptone, and 0.5% NaCl). Bacterial growth was evaluated considering the following intervals: 0, 2, 4, 6, 8, 10, 12, 24, 30, 36, and 48h. After incubation at 35°C for 18h, the viable cells were counted in plate count agar and expressed as CFU per milliliter. The experiment was performed in triplicate. Further, the biofilm production of N–341 was evaluated considering the intervals determined in the bacterial growth curve.
Preparation of biofilm samples for SEMBacterial growth was observed on Petri dishes containing glass cover slips for biofilm adhesion. The isolates N–354, N–365, and N–341 were cultivated in TSA with 0.24% glucose overnight, adjusted to the 0.5 McFarland scale, and diluted 1:10 in TSA with 0.24% of glucose. The aliquots (2mL each) were placed in each Petri dish containing three glass coverslips and were statically incubated at 35°C for 4, 8, 12, and 24h. After incubation, the Petri dish was washed three times with saline (0.85% NaCl) to remove all planktonic cells. The adherent cells were fixed with 5% glutaraldehyde for 5h. After fixation, the plate was washed three times with 0.1M sodium cacodylate buffer. For SEM, S. aureus cells were fixed for 30min at room temperature with 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer (pH 7.2) and post-fixed for 30min at room temperature with 1% OsO4 solution containing 2.5mM CaCl2 in the same buffer. The cells were dehydrated in an ascending acetone series and dried using the critical point method with CO2 (CPD 030, Balzers, Switzerland). Subsequently, the samples were mounted on aluminum stubs, coated with a 20-nm gold layer, and examined under a scanning electron microscope (Jeol JSM6390LV) at the Rudolf Barth Electron Microscopy Platform of Institute Oswaldo Cruz.
Evaluation of pheno-genotypic resistance to beta-lactamicAll the 20 S. aureus isolates were subjected to disk diffusion tests using cefoxitin (30μg), oxacillin (10μg), penicillin (10 UI), and amoxicillin+clavulanic acid (30μg) disks. In addition, the “edge zone” test was used to evaluate the production of beta-lactamases. After overnight incubation at 35°C, followed by inhibition zone measurement,16,17 the results were evaluated as per the interpretation criteria following the CLSI standards.15 PCR was performed for mecA using primers designed by Murakami et al.24 and Melo et al.25 and for blaZ according to Rosato et al.26 (Table 1).
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) tests were performed according to CLSI15 for the isolates N–354, N–365, and N–341 using different concentrations ranging from 0.25, 0.5, 1.0, 2.0, 4.0, 8.0, 16, 32, 64, 128, 256 to 512μg/mL in MH broth. For MBC determination, cefoxitin cultures with concentrations above the MIC were inoculated in AMH.27
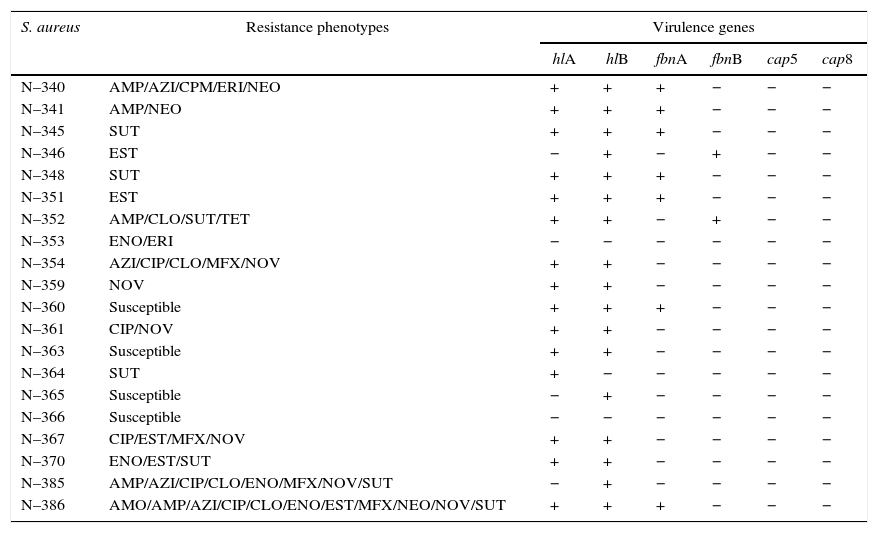
Results and discussionThe S. aureus isolates, which were identified by phenol-genotypic and proteomic techniques, were tested for sensitivity to certain antimicrobial agents and the presence of virulence genes for the fibronectin, hemolysin, and capsule. These results were used to create profiles highlighting the more distinct characteristics of the strains and representatives were collected. Therefore, 20 S. aureus strains were selected considering their antibiotic resistance profiles and the presence of virulence genes (Table 2).
Antibiotyping profile and virulence genes of the 20 S. aureus strains included in this study.
| S. aureus | Resistance phenotypes | Virulence genes | |||||
|---|---|---|---|---|---|---|---|
| hlA | hlB | fbnA | fbnB | cap5 | cap8 | ||
| N–340 | AMP/AZI/CPM/ERI/NEO | + | + | + | − | − | − |
| N–341 | AMP/NEO | + | + | + | − | − | − |
| N–345 | SUT | + | + | + | − | − | − |
| N–346 | EST | − | + | − | + | − | − |
| N–348 | SUT | + | + | + | − | − | − |
| N–351 | EST | + | + | + | − | − | − |
| N–352 | AMP/CLO/SUT/TET | + | + | − | + | − | − |
| N–353 | ENO/ERI | − | − | − | − | − | − |
| N–354 | AZI/CIP/CLO/MFX/NOV | + | + | − | − | − | − |
| N–359 | NOV | + | + | − | − | − | − |
| N–360 | Susceptible | + | + | + | − | − | − |
| N–361 | CIP/NOV | + | + | − | − | − | − |
| N–363 | Susceptible | + | + | − | − | − | − |
| N–364 | SUT | + | − | − | − | − | − |
| N–365 | Susceptible | − | + | − | − | − | − |
| N–366 | Susceptible | − | − | − | − | − | − |
| N–367 | CIP/EST/MFX/NOV | + | + | − | − | − | − |
| N–370 | ENO/EST/SUT | + | + | − | − | − | − |
| N–385 | AMP/AZI/CIP/CLO/ENO/MFX/NOV/SUT | − | + | − | − | − | − |
| N–386 | AMO/AMP/AZI/CIP/CLO/ENO/EST/MFX/NEO/NOV/SUT | + | + | + | − | − | − |
AMO, amoxicillin; AMP, ampicillin; AZI, azithromycin; CIP, ciprofloxacin; CLO, chloramphenicol; CPM, cefepime; ENO, enrofloxacin; ERI, erythromycin; EST, streptomycin; MFX, moxifloxacin; NEO, neomycin; NOV, novobiocin; SUT, cotrimoxazole; TET, tetracycline. +, positive amplification; −, negative amplification.
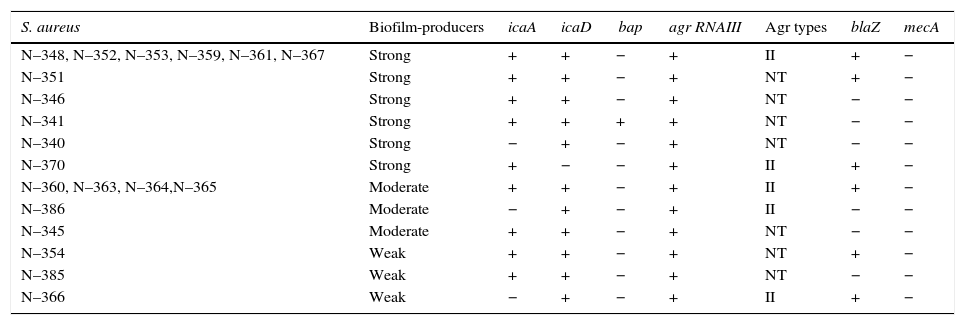
All the 20 S. aureus strains were biofilm producers, classified as strong (55%=11/20), moderate (30%=6/20), and weak (15%=3/20) producers. It is important to detect Staphylococcus spp. Isolates that produce biofilms because this virulence factor guarantees the installation and maintenance of the bacteria in the glandular breast tissue.28 Moreover, the polysaccharide mucus of biofilms facilitates bacterial adhesion to biomaterials, which is not removable despite repeated washings; therefore, its production is associated with infections caused by milking machines.29icaA and icaD were detected in 17 (85%) and 19 (95%) isolates, respectively. Sixteen isolates tested positive for both the genes and only one (5%) tested positive for bap (Table 3). The low incidence of bap indicates that the ica-dependent mechanism, slime producer, may be primarily responsible for the adhesion and biofilm formation in the strains, as reported by Vautor et al.30 All the biofilm-producing strains presented either icaA or icaD or both. However, no relation was observed between the biofilm formation intensity and number of amplified icaA and icaD. Arciola et al.31 did not observe any relationship between slime production and the presence of these genes, suggesting that it could be a consequence of the experimental conditions such as lower sugar concentration in the agar or shorter incubation period.
Biofilm production, agr system classification and presence of the genes icaA, icaD, bap, agr RNA III, blaZ and mecA in S. aureus strains isolated from bovine mastitis.
| S. aureus | Biofilm-producers | icaA | icaD | bap | agr RNAIII | Agr types | blaZ | mecA |
|---|---|---|---|---|---|---|---|---|
| N–348, N–352, N–353, N–359, N–361, N–367 | Strong | + | + | − | + | II | + | − |
| N–351 | Strong | + | + | − | + | NT | + | − |
| N–346 | Strong | + | + | − | + | NT | − | − |
| N–341 | Strong | + | + | + | + | NT | − | − |
| N–340 | Strong | − | + | − | + | NT | − | − |
| N–370 | Strong | + | − | − | + | II | + | − |
| N–360, N–363, N–364,N–365 | Moderate | + | + | − | + | II | + | − |
| N–386 | Moderate | − | + | − | + | II | − | − |
| N–345 | Moderate | + | + | − | + | NT | − | − |
| N–354 | Weak | + | + | − | + | NT | + | − |
| N–385 | Weak | + | + | − | + | NT | − | − |
| N–366 | Weak | − | + | − | + | II | + | − |
NT, not typable; +, positive; −, negative.
All the S. aureus isolates tested positive for agr RNAIII. Although RNAIII is not essential for S. aureus growth in vitro, it modulates the expression of genes involved in its pathogenesis.32 In fact, the roles of agr and quorum sensing in biofilm formation remain elusive. The Agr type II group was prevalent in 14 isolates, but the remaining six isolates could not be typed by the adopted technique. Moreover, Melchior et al.33 found a high prevalence of Agr type II in 81% S. aureus isolates from bovine mastitis in the Netherlands, whereas 9% were Agr type I group. In addition, they suggested Agr type II strains are better adapted to the dairy environment than Agr type I strains. A study by Fabre-Klein et al.34 suggested that S. aureus strains increase biofilm production to adapt to the milk-filled environment of the udder. The absence of the agr system function facilitates the initial adhesion of staphylococci to surfaces, presumably due to the positive expression of adhesion molecules and negative expression of biofilm formation factors, which are released in the late stationary phase.35
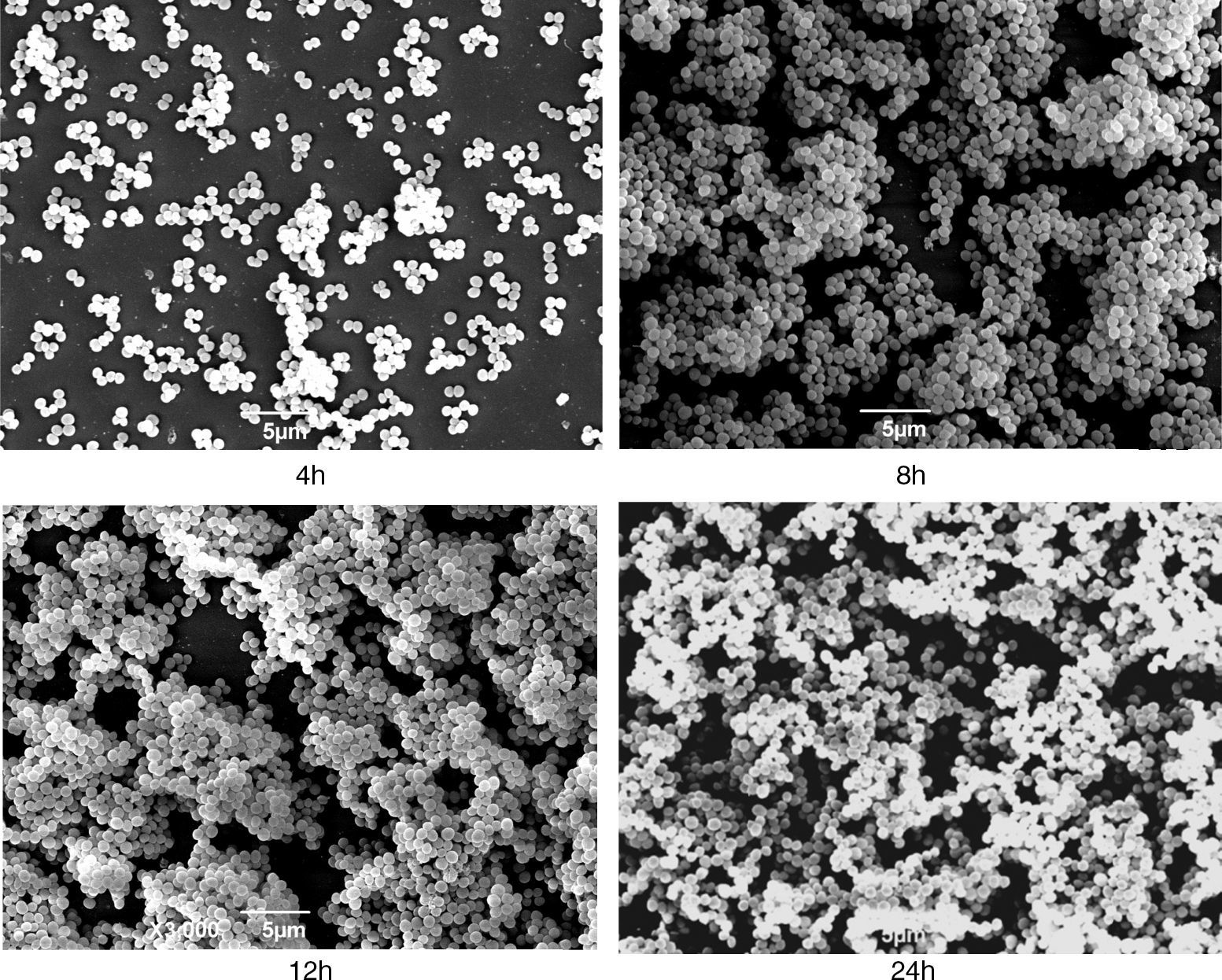
The S. aureus isolate N–341was tested positive for icaA, icaD, and agr and presented a strong biofilm production in the assays; therefore, it was selected for the growth curve estimation. The biofilm production of N–341 in microplates was evaluated according to the time intervals of the growth curve assay considering the following phases: lag until 4h, exponential reaching the plateau at 12h, and death from 24h. The biofilm production reached its peak at 12h (OD=0.596). At SEM, this isolate displayed a meshwork-like structure associated with the surface and with various gaps at 8h of growth; this was not apparent at 4h. At 12h of growth, these gaps decreased in size and the cell layer became thicker, indicating the possible establishment of the biofilm. Within 24h, the surface was filled with dense cell clusters (Fig. 1), probably indicating the next stage of biofilm formation. Using this model, it was possible to detect gradual changes in the biofilm complexity during the different stages of S. aureus growth. Therefore, our model used for biofilm formation and the procedure for cultivation of strains in TSA with 0.24% glucose in microplates and glass cover slips for 24h at 35°C without agitation can be satisfactorily used.
Scanning electron micrographs of strain N–341 showing morphological changes associated with growth. Meshwork-like structures associated to the surface with various gaps were observed at 8h of growth, these structures were not apparent at 4h. At 12h of growth, the gaps decreased in size and the cell layer became denser and at 24h the surface was filled with dense cell clusters.
Recently, Savage et al.36 reported that biofilm mode of growth increases horizontal transfer of plasmid-borne antibiotic resistance determinants by conjugation in S. aureus. In addition, the biofilm can act as a barrier preventing the adsorption/penetration of antimicrobials, and the matrix promotes their dilution to subinhibitory concentrations. Moreover, the difference in bacterial physiology presented in the biofilm can influence the efficacy of antibiotics.37 In this study, all isolates were sensitive to oxacillin and cefoxitin in the disk diffusion test, supporting the results of previous studies stating that the prevalence of oxacillin resistance in S. aureus isolates from bovine mastitis is low.38,39 In the diffusion disk test, 15 isolates showed a sensitivity zone >29mm, but five were resistant to penicillin. The CLSI indicates that staphylococci producing beta-lactamase may be phenotypically sensitive; therefore, before reporting their sensitivity, it is recommended that these isolates should be tested for beta-lactamase production. The recommended phenotypic test for the production of beta-lactamase in S. aureus, i.e., the edge zone test, interprets the growth on the border of the inhibition zone. This test is considered more sensitive for S. aureus than the nitrocefin test.16 In the edge zone test, 100% (15/15) of the sensitive isolates were positive and were reported as penicillin resistant. A previous study has reported high resistance to penicillin in coagulase-positive staphylococci isolated from bovine mastitis cases.38 However, this antibiotic is rarely considered a treatment option for mastitis, although some producers still insist on using it due to its low cost.
Furthermore, the susceptibility to amoxicillin associated with the beta-lactamase inhibitor was 100%, suggesting that the mechanism of beta-lactamase production may lead to penicillin resistance. Among the isolates considered resistant to penicillin, 70% (14/20) possessed blaZ and were tested negative for mecA using two different primers. Of the six isolates tested negative for both blaZ and mecA, five displayed moderate or strong biofilm production.
The MIC and MBC for cefoxitin were analyzed for the isolates N–354, N–365, and N–341. The stronger biofilm producer N–341 presented the highest MIC and MBC (1and 64μg/mL, respectively), followed by the moderate producer N–365 (<0.25and 4μg/mL, respectively). The weaker biofilm producer N–354 presented the lowest MIC and MBC (<0.25μg/mL for both). Since N–341 was negative for blaZ and mecA, the high cefoxitin MBC value may be associated with biofilm protection. Wells et al.40 discussed that there is no universal acceptable methodology for evaluating antimicrobial resistance and biofilm production. Based on this, it appears that exopolysaccharides (EPS) secreted by the bacteria act as a barrier that may play a role in this resistance, preventing the adsorption and penetration of antimicrobials. Moreover, the EPS matrix could neutralize or bind these compounds, promoting their dilution to subinhibitory concentrations before they reach the cells. In addition, biofilms are composed of both dormant and active cell subpopulations. This difference in the bacterial physiology can influence the efficacy of antibiotics.37
In conclusion, all the 20 isolates were biofilm producers and mostly presented icaA and icaD; only the N–341 strain was positive for bap. agr RNAIII was detected in all the isolates, and Agr type II showed a significant prevalence. The N–341 strain showed gradual changes in the complexity of the biofilm along the phases of growth curve in SEM, reaching the peak at the stationary phase. Moreover, the detected penicillin resistance was related to the production of beta-lactamase due to the absence of mecA and sensitivity to amoxicillin+clavulanic acid in all the isolates. Finally, because N–341 was negative for the tested resistance genes, the cefoxitin MBC of 64μg/mL may be associated with biofilm protection since this strain is a strong producer of this virulence factor.
Taken together, these data suggest that a greater understanding of biofilm formation may add to our knowledge on bacterial resistance in vivo. Thus, studies that uncover colonization factors in biofilm formation are important and will form the basis for the development of treatments for bacterial resistance in biofilms.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by the National Council for Scientific and Technological Development (CNPq, Rio de Janeiro, Brazil – process 308528/2011–5), Foundation for Research Support in the State of Rio de Janeiro (FAPERJ; process E–26/112.658/2012) and Coordination for the Improvement of Higher Education Personal (CAPES).