Avian pathogenic Escherichia coli (APEC) isolates from apparently healthy free range helmeted guineafowl were characterized. Most of them had a high frequency of virulence associated genes, multi drug resistance and high pathogenicity. We demonstrated that helmeted guineafowl have potential to transmit antibiotic resistant APEC to other species including humans.
The Avian Pathogenic Escherichia coli (APEC) strains are responsible for a large number of extra-intestinal diseases in birds, either locally or as systemic infections; those are the so called avian colibacillosis and are responsible for economic losses for the world's poultry industries.1 In addition, APEC strains are a subpathotype of pathogenic extra-intestinal bacteria (ExPEC), category responsible for diseases in humans and animals such as urinary tract infection (UTI), neonatal meningitis and septicemia,2 suggesting that APEC strains may be a potential zoonotic pathogens.3
Healthy birds can eliminate strains that harbors virulence genes to the environment, thus becoming a sources of infection to other animals and to humans.4,5 In this regard, the increase of drug resistant strains, due to the indiscriminate use of antibiotics in animal production, can transfer resistance genes to other bacteria of the environment and to normal human microbiota.6–8
There is no consensus in the literature to define the APEC pathotype and the development of methodologies for its diagnosis are fault; mainly due to it be a very heterogeneous group of microorganisms, in which different isolates can harbor a different associations of virulence factors (VFs), each capable of inducing avian colibacillosis.9 Also, free-range chickens and human often share the same environment, in this sense, these birds and their products (meat and eggs) can act as ExPEC sources of infection to humans.10 Like any source of animal protein, helmeted guineafowl may act as a possible carrier of pathogenic microorganisms to humans; however, until now, no studies focused on this species as been possible carriers. Thus, we performed this study to detect and characterize drug-resistant pathogenic E. coli carrying virulence genes related to APEC pathotype in free-range helmeted guineafowl and clarified how these birds can be relevant in humans and other animal's infections.
This study was approved by the Committee on Ethics for the Use of Animals of (CEUA), protocol number 19.008/16. Samples were collected from 56 free range helmeted guineafowl without history of the use of antibiotics, of unknown genetic origin and of different age groups from 4 small farms in Jaboticabal city, Sao Paulo State, Brazil. In the first farm 6 samples was collected, 12 samples in the second farm, 48 samples in the third farm and 46 in the fourth farm, totalizing 56 samples from the cloaca and 56 from the oropharynges that were obtained with the aid of sterile swabs, which were placed in sterile tubes, containing 5mL of BHI broth (Brain Heart Infusion) and kept in under refrigeration until processed.
From the swabs inoculated into BHI and incubated at 37°C for 16h, a Polymerase Chain Reaction (PCR) screening was performed for the detection of the cvaC, iroN, iss, iutA, ompT and hlyF genes. Protocols for the DNA template preparation, PCR and primer oligonucleotides for these genes were taken from the EcL protocol, available at http://www.apzec.ca/en/APZEC/Protocols/APZEC_PCR_en.aspx. Then, samples that were positive for at least five of the above cited genes were used to detect E. coli isolates.11,12 In addition, obtained isolates were subjected to a new PCRs to detect the following additional 11 virulence genes: sitA, tsh, traT, vat, astA, iucC, iucD, papC, irp2, fimH and fyuA. The frequencies of VAGs were compared with Fisher's exact test using Prism for Windows version 6.01 (GraphPad Software). Associations were considered statistically significant if the calculated P-value was <0.05.
E. coli phylogroups were typing by PCR phylotyping method of Clermont et al. (2013)13 targeting chuA and yjaA genes and TspE4.C2 DNA fragment. Serotyping of the APEC strains were performed using somatic (O1-O181) and flagellar (H1-H56) antigens that were produced at the Bacteriology Center of the Adolfo Lutz Institute – Sao Paulo, Brazil. Additionally, the isolates antimicrobial susceptibility was performed by disc diffusion method.14 Antimicrobials tested were: ampicillin (amp) (10μg), cephalothin (cep) (30μg), streptomycin (str) (10μg), gentamicin (gen) (10μg), ciprofloxacin (cip) (5μg), chloramphenicol (chl) (30μg), tetracycline (tet) (30μg), nitrofurantoin (nit) (300μg), sulfamethoxazole+trimethoprim (sut) (25μg), ceftiofur (ctf) (30μg), ceftriaxone (cro) (30μg), amikacin (ami) (30μg), cefoxitin (cfo) (30μg), kanamycin (kan) (30μg), amoxicillin+clavulanic acid (amc) (30μg), norfloxacin (nor) (10μg) and fosfomycin (fos) (50μg). E. coli isolates were also screened for extended-spectrum beta-lactamase (ESBL) genes for blaCTX-M genotype groups 1,15 216 and 9,17blaTEM and blaSHV.18,19
Pathogenicity test was determined according to Guastalli et al. (2013).20 Bacterial culture (0.1ml) was inoculated into the left thoracic air sac of day-old chicks. For inoculum preparation, a colony of each bacterial strain was seeded in 10ml of BHI broth, incubated for 18h at 37°C and subsequently diluted to a 1:10 ratio. Inoculum concentration was standardized to 107CFU/mL. The E. coli (serogroup O1) belonging the Laboratory of Ornithopathology of USP, was used as a positive control. Negative control birds were inoculated with BHI broth only. For each strain, as well as for the negative and positive control groups, ten male chicks from a commercial lineage were used. The strains were classified due to its mortality as follow: high (≥80%), intermediate (>50% and <80%), low pathogenicity (≤50%) and non-pathogenic (zero mortality).
Genomic DNA digestion with XbaI (Invitrogen, USA) was performed as described by Ribot et al. (2006)21 with modifications and a Salmonella Braenderup H9812 strain was used as a molecular weight reference. The electrophoresis occurred at 14°C, on a 1% Pulsifield Certified agarose gel, with initial time of 2.2s, final time of 54.2s, on a gradient of 6Vcmx−1, an angle of 120°, for 23h. The fragments similarity was compared using the Dice coefficient at 1% tolerance and 0.5% optimization and the dendrogram was calculated using the neighbor joining method with BioNumerics software version 7.1 (Applied Maths, Sint-Martens-Latem, Belgium). The housekeeping genes adk, fumC, gyrB, icd, mdh, purA and recA were amplified by PCR for the MSLT analysis. These genes were chosen based on the MLST protocol for E. coli from the University of Warwick, USA.22 The specific oligonucleotide as well as the PCR amplification conditions are available at http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/documents/primersColi_html. The PCR products were sequenced in an ABI 3100 sequencer (Applied Biosystems, Waltham, USA) with Big Dye Terminator v3.1 kit (Applied Biosystems, Waltham, USA). The resulting sequences were analyzed by the Phred/Phrap/Consed program package.23–25
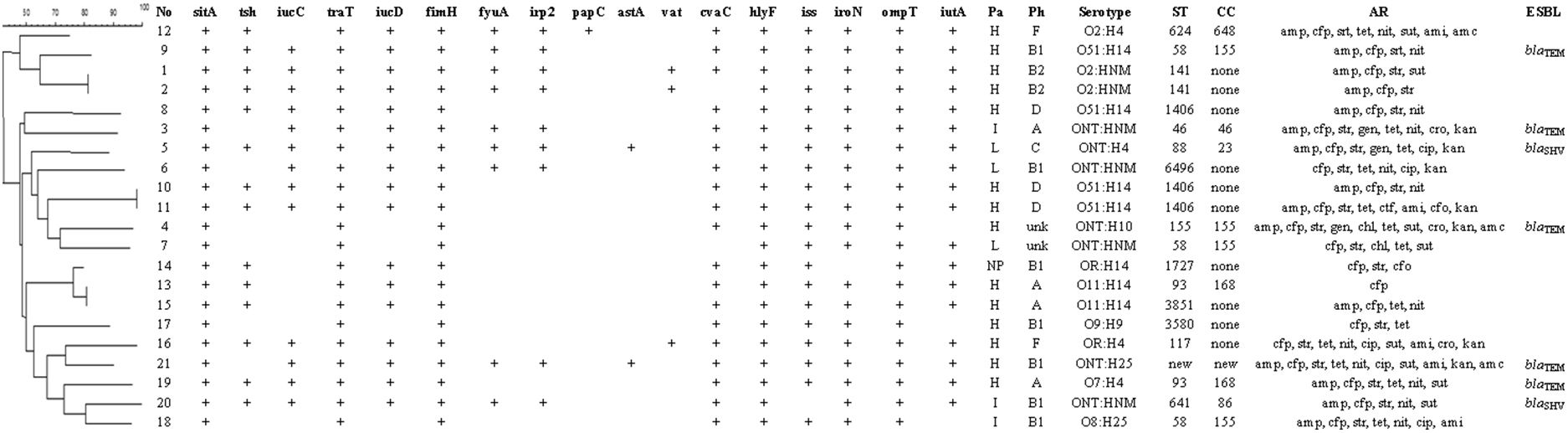
All the results are shown in Fig. 1. From the 112 samples, 21 isolates were obtained, 20 were from cloaca and one from the oropharynx. These isolates were positive for at least five of the APEC related genes (cvaC, hlyF, iss, iroN, ompT and iutA) and were, further, subjected to another PCR for the detection of additional virulence genes. Isolated E. coli gene profile is described at Table 1.
Association between dendrogram analysis of genetic diversity by PFGE and virulence indicators of the 21 APEC isolates. No: isolate number; Pa: pathogenicity; Ph: philogeny; H: high; I: intermediate; NP: non-pathogenic; L: low; unk: unknown; ST: stypes; CC: clonal complex, AR: antimicrobial resistance; amp: ampicillin; cep: cephalothin; str: streptomycin; gen: gentamicin; cip: ciprofloxacin; chl: chloramphenicol; tet: tetracycline; nit: nitrofurantoin; sut: sulfamethoxazole+trimethoprim; ctf: ceftiofur; cro: ceftriaxone; ami: amikacin; cfo: cefoxitin; kan: kanamycin; amc: amoxicillin+clavulanic acid; nor: norfloxacin; fos: fosfomycin; ESBL: extended-spectrum beta-lactamase genes.
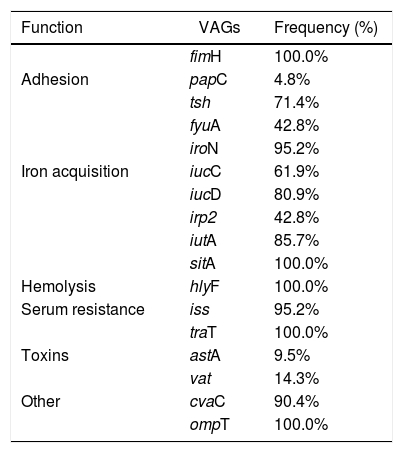
Frequency correlation of each virulence-associated gene (VAGs) in the 21 potentially APEC isolates.
| Function | VAGs | Frequency (%) |
|---|---|---|
| fimH | 100.0% | |
| Adhesion | papC | 4.8% |
| tsh | 71.4% | |
| fyuA | 42.8% | |
| iroN | 95.2% | |
| Iron acquisition | iucC | 61.9% |
| iucD | 80.9% | |
| irp2 | 42.8% | |
| iutA | 85.7% | |
| sitA | 100.0% | |
| Hemolysis | hlyF | 100.0% |
| Serum resistance | iss | 95.2% |
| traT | 100.0% | |
| Toxins | astA | 9.5% |
| vat | 14.3% | |
| Other | cvaC | 90.4% |
| ompT | 100.0% |
Phylogenetic analysis revealed that most of the isolates (33.3%) belong to group B1, followed by groups A (19.0%), D (14.3%), B2 (9.5%), F (9.5%) and C (4.8%) and that two isolates (4 and 7) could not be typed. Isolates belonging to groups B2 and F were associated with a higher number of virulence factors, with a mean of 14.5 and 13.5 per isolate of each group, respectively.
A single isolate showed resistance to only one antimicrobial. The remaining 20 isolates were resistance to at least three antimicrobial simultaneously and 19 of these isolates were resistant to three or more antimicrobial classes, which represent 90.4% of multi drug resistant. The highest drug resistance was found against cephalotin (100.0%), streptomycin (90.5%), ampicillin (71.4%) and tetracycline (61.9%). As for the antimicrobial classes, 42.8% of the isolates were resistant to penicillins, 32.2% to cephalosporins and 40.5% to aminoglycosides. ESBL genes for blaTEM were found in resistant isolates 3, 4, 9, 19 and 21 and blaSHV in isolates 5 and 20.
Thirteen of the 21 samples analyzed were O antigen typable (61.9%) and were within the following six serogroups: O2 (14.3%), O51 (19.0%), O11 (9.5%), O7 (3.2%), O8 (3.2%) and O9 (3.2%). Of the remaining strains, seven (33.3%) were non typable (NT) and two (9.5%) could not be characterized due to its rough-looking colonies (RL). The H antigen could not be determined in six samples (28.5%) due to they been immobile (IS). The other 16 (76.2%) strains were typable and five antigens were identified: H4 (28.5%), H14 (23.8%), H25 (9.5%), H9 (3.2%) and H10 (3.2%). The most prevalent O:H serotypes were: O51:H14 (19.0%) and NT:IS (19.0%)
The pathogenicity test revealed that 14 (66.6%) strains were highly pathogenic (HP), three (14.8%) strains were intermediate pathogenic (MP), three (14.8%) strains were low pathogenic (LP) and one (4.7%) was a nonpathogenic (NP) strain. It was observed that all of the birds in the positive control group died, while all birds of the negative control group lived. The clinical signs and macroscopic lesions were observed with higher frequency with high and intermediate pathogenic strains. None of the VAGs were statistically significant to differentiated HP/MP from LP/NP strains based Fisher's test.
The dendogram showed two large clusters with three subdivisions and the 21 possible APEC isolates generated 18 different pulsotypes, of which only three were shared, those been: 1 and 2, 10 and 11, 13 and 15. Sixteen sequence types (ST) were identified as follow: ST 624, ST 58, ST 1406, ST 46, ST 88, ST 6496, ST 155, ST 1727, ST 93, ST 3851, ST 3580, ST 117, ST 83, ST 641 and a new one (ST) that corresponded to the isolate 21.
There are no data about helmeted guineafowl potential as sources of APEC infection for other animals. Moreover APEC virulence mechanisms are not yet fully understood and it is known that APEC strains have different virulence profiles that correlate to the animal species and the region in which it was isolated.12,26,27 In several studies authors have shown that APEC-related genes iutA, hlyF, iss, iron, ompT and cvaC occur more frequently and are associated with highly pathogenic strains.12,28,29 All 21 isolates showed at least five of these and others important genes associates with ExPEC that have been detected in other animals like pigeons30 and humans.31 These findings suggest that wild and domestic bird may act as sources of human pathogenic E. coli, thus reinforcing its zoonotic nature.3
Most of the isolates were found highly pathogenic by the pathogenicity test, with low and intermediate pathogenic strains composing only 29.6% and only one isolate was nonpathogenic. The high number of high pathogenic isolates in apparently healthy birds can be attributed to the fact that these genes were not expressed because according to Won et al. (2009),32 the pathogenicity of APEC is based not only on the gene presence, but also on its expression. In addition, APEC infections are multifactorial and highly dependent on the microorganism/host interactions, which develop secondarily to other factors, such as poor environmental and handling conditions and pre-existent infections that can affect the host immune system or the respiratory epithelium.32,33 It is also worth mentioning that a significant genetic material exchange between bacteria may occur, making possible that horizontal gene transfer from pathogenic to non-pathogenic strain lead to the emergence of new virulent strains between the normal human microbiota.7
Of today concern is the fact that a large number of E. coli strains causing human infections, including those resistant to antimicrobials, are of animal origin.5,34 According to Mellata et al. (2013),35E. coli samples isolated from birds are commonly resistant to more than one antimicrobial. This profile was also over served here with 95.2% of the isolates exhibiting resistance to at least three antimicrobials, fact similar to others studies.36,37 The highest drug resistance was observed against cephalothin, streptomycin, ampicillin and tetracycline which are the most commonly used antibiotics in human infections. In addition, finding ESBL genes in these animals is a concern because transmission of ESBL isolates between humans and other species has been reported.38
Phylogenetic analysis revealed that most of the isolates from groups B1, and B2 and F strains have a highest number of VAGs. Although recent studies that utilize the updated method by Clermont et al. (2013)13 are scarce, Logue et al. (2017)39 classified APEC isolates according to the new phylogenetic typing and concluded that strains in A and B1 group were of lower pathogenic potential. In contrast, in this study, three samples from group A were classified as high pathogenic and one as intermediate. The only non-pathogenic strain belonged to the B1 group, as well as a low pathogenicity strain and, interestingly, two intermediate pathogenicity strains and three high pathogenic strains were included in this group. Thus, it was noticed that strains of low and high pathogenic potential can be of the same phylogenetic group and more studies are needed in order to clarify, effectively, the phylogeny relationship of APEC isolates.
Strains involved in colibacillosis are often associated with three major serogroups: O1, O2 and O78, with the other ones been sporadic, but also occurring.3,40 In this study, 14.3% of the isolated belonged to the O2 group, which is in agreement with Schouler et al. (2012),9 whose study found out that this serogroup harbored mainly pathogenic samples. In our findings, all O2 samples were pathogenic. In addition, the serogroup H4 was the most commonly found, with the O51:H14 serotype has been the most prevalent.
PFGE analysis revealed a great intra-specific variability. Among the isolates that shared the same pulsotype, samples 1 and 2, from the same animal, have genetic similarities and shared the other evaluated characteristics but differ among themselves in the presence of the cvaC. Isolates 13 and 15, which were from the same animal, had a similar genotypic profile, pathogenicity and phylogeny, but they had differences in others parameters. Samples 10 and 11, isolated from different birds, were similar in all characteristics and, possibly, there are bacterial clones. These results that can be explained by the fact that these birds share the same environment and were in close proximity, facilitating genetic material exchange,7 fact that suggesting the possibility of free-range helmeted guineafowl to transmit potential APEC strains to other animals including humans.
Sixteen sequence types (ST) were identified of 21 isolates, a new ST, corresponding to isolate 21, which differed in only one locus from the ST 155 (data not shown). There was similarity among isolates which share the same ST as well as between isolates with different STs. In general, phylogenetic analysis by MLST has shown that, in fact, APEC constitutes a heterogeneous group, fact that was also observed in another study.41
Maluta et al. (2014)29 verified that APEC and ExPEC in humans can shared ST 155, ST 88 and ST 117, which were found in free-range helmeted guineafowl, with the ST 117, being the most common among isolates related to intensive care units. The ST 46 and ST 83, also associated with humans and other animals like dogs42 and cats43 was found in this study too. Helmetd guineafowl APEC isolates sharing the same phylogenetic background with ExPec strains proves that these animals can be a source of infection in humans and other animals, include pet companions.
The diversity of factors combinations that may lead to virulence and to antimicrobials resistance reveals the great importance of APEC zoonotic potential. Our data showed that healthy free-range helmeted guineafowl are an underestimated how natural reservoirs of APEC with high pathogenic potential and multi drug resistance. This is aggravated because of the fact that these bacteria can be eliminated by the respiratory tract and by feces, thus making these birds a sources of infection of ExPEC to other animals, including humans.
The authors would like to thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP – [grant number 2014/06313-3] and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for all research support granted.