Freezing temperatures are a major challenge for life at the poles. Decreased membrane fluidity, uninvited secondary structure formation in nucleic acids, and protein cold-denaturation all occur at cold temperatures. Organisms adapted to polar regions possess distinct mechanisms that enable them to survive in extremely cold environments. Among the cold-induced proteins, cold shock protein (Csp) family proteins are the most prominent. A gene coding for a Csp-family protein, cspB, was cloned from an arctic bacterium, Polaribacter irgensii KOPRI 22228, and overexpression of cspB greatly increased the freeze-survival rates of Escherichia coli hosts, to a greater level than any previously reported Csp. It also suppressed the cold-sensitivity of an E. coli csp-quadruple deletion strain, BX04. Sequence analysis showed that this protein consists of a unique domain at its N-terminal end and a well conserved cold shock domain at its C-terminal end. The most common mechanism of Csp function in cold adaption is melting of the secondary structures in RNA and DNA molecules, thus facilitating transcription and translation at low temperatures. P. irgensii CspB bound to oligo(dT)-cellulose resins, suggesting single-stranded nucleic acid-binding activity. The unprecedented level of freeze-tolerance conferred by P. irgensii CspB suggests a crucial role for this protein in survival in polar environments.
Living organisms encounter several serious challenges when they are exposed to cold environments, including reduced enzyme activity, decreased membrane fluidity, reduced transport of nutrients and waste products, reduced protein movement, decreased rates of transcription and translation, slow DNA replication, retarded protein folding, cold-denaturation of proteins, and intracellular ice formation.1–3 Although freezing temperatures are a major environmental threat at the North and South Poles, relatively high numbers of diverse microorganisms thrive in the polar regions. In particular, α-, β-, and γ-proteobacteria and the Cytophaga-Flavobacterium-Bacteroides phyla are the most commonly found bacteria, and eukaryotes, such as yeasts and microalgae, are frequently found in polar environments.4 Organisms living in the polar regions must have the ability to adapt to extremely cold temperatures. To avoid the transition from a fluid- to gel-phase cell membrane, the proportion of unsaturated and methyl-branched fatty acids in lipid membranes is increased.5 The formation of stable secondary structures in nucleic acids at low temperatures inhibits replication, transcription, and translation. Thus, nucleic acid-binding proteins, such as Csp-family proteins and RNA helicases, are induced during temperature downshifts and function as RNA chaperones,6 melting the stable secondary structures in RNA molecules, which facilitates translation and ribosome biogenesis.7–9 Several chaperones, such as GroEL10 and DnaK,11 are induced upon cold shock, possibly to cope with cold-denatured proteins. Protein folding, especially cis/trans isomerization of peptidyl prolyl bonds, occurs very slowly at low temperatures, and peptidyl prolyl isomerases play an important role in cold adaptation by facilitating the folding of functionally significant proteins.12 Cryoprotectants, such as antifreeze proteins,13 trehalose,2 and exopolysaccharides,14 have also been implicated in protection of polar organisms, either by preventing ice crystal formation or by avoiding dehydration. Genomic sequencing results suggest some other characteristics of psychrophiles, like enhanced antioxidant capacity to cope with increased production of toxic reactive oxygen species.15
Increasing numbers of Csp homologs are being reported from psychrophiles, mostly through genomic DNA sequencing projects, but only limited functional data are available. In an effort to elucidate the detailed mechanisms that allow polar organisms to survive at low temperatures, we studied the functional roles of Csp-family proteins in polar bacteria. A psychrophilic bacterium, Polaribacter irgensii KOPRI 22228, was isolated from Arctic Sea sediment.16 Two csp genes, cspAPi and cspCPi, were previously cloned from this bacterium and exhibited considerable homology to canonical Escherichia coli cspA (CspAEc). Overexpression of either of these two genes conferred significant cold-resistance phenotypes to their recombinant hosts, and also complemented the cold-sensitivity of a quadruple csp deletion mutant BX04 (ΔcspA, ΔcspB, ΔcspG, and ΔcspE) strain,17 suggesting functional homology among those Csp-family proteins.16 In this study, we report another Csp-homologous protein from P. irgensii KOPRI 22228, CspBPi, which contains an extra domain on its N-terminal region, in addition to the well conserved cold-shock domain (CSD) at its C-terminal region. This belongs to a subfamily of CSD-fold proteins, recently identified through metagenomic studies of psychrophilic bacteria, and no functional data is available for this kind of proteins at our best knowledge. Therefore, the function of this unique Csp protein from an arctic bacterium in conferring cold tolerance was analyzed in this study.
Materials and methodsBacterial culture and cloning of the cspBPi geneP. irgensii KOPRI 22228 was isolated from Arctic Sea sediments near Dasan Korean Arctic Station (Ny-Alesund, Norway), and cultured as previously reported.16 The cspBPi gene (GenBank WP_004570868) was obtained by PCR amplification. The template DNA was extracted from P. irgensii KOPRI 22228 cells using G-spin™ bacteria genomic DNA extraction kit (Intron Co., Korea), according to the protocol suggested by the manufacturer. The forward primer sequence was CspBPi F1H, 5′-AGTAAGCTTATGGCAAAATCGCAGCAGACCT-3′, and the reverse primer was CspBPi B150*B, 5′-CCGGATCCTTATATTTTGGTAACTTTAACTGCATTCATT-3′ (manufactured by Bioneer Co., Daejion, Korea). The PCR mixture consisted of 5μl of 10× PCR buffer (final concentrations: 50mM KCl, 0.01% gelatin, 10mM Tris–HCl, pH 9.0), 2.5mM MgCl2, 0.2mM of each dNTP, 200nM of each primer, 1μl of template DNA, and 2.5 units of Taq DNA polymerase (Takara, Japan) in the final 50μl volume. The PCR was performed in a DNAEngine thermal cycler (Bio-Rad Laboratories Inc., USA) using a cycling condition that consisted of an initial denaturation at 95°C for 5min and then 30 cycles with denaturation at 94°C for 1min, annealing at 55°C for 30s, and extension at 72°C for 1min. A final extension was performed at 72°C for 5min. The PCR products that were 0.5kb in size were double-digested with HindIII and BamHI, and cloned using pAED4,18 an E. coli expression vector, digested with the same enzymes. To avoid any mutations arising from error-prone Taq DNA polymerase reactions, several clones were picked for sequencing analysis. The resulting plasmids was named pAED-cspBPi.
Expression of CspBPi in E. coliThe recombinant plasmid for expression of CspBPi, pAED-cspBPi, was transformed into competent E. coli BL21(DE3) cells (Invitrogen Co., CA, USA) using the method described by Sambrook et al.19 For overproduction of CspBPi, 1ml of the overnight liquid culture was transferred to a flask containing 50ml fresh LB medium containing 100μg/ml ampicillin. When cell cultures had reached an OD600 of 0.4, IPTG was added to the final concentration of 0.1mM. The cultures were incubated further at 37°C for 2h with vigorous shaking. Overexpression of the CspBPi was analyzed by 20% SDS-PAGE. The protein bands were visualized by Coomassie brilliant blue R250 staining.
Resistance to freezing and thawingCspBPi protein was overexpressed in E. coli as described above. As the experimental control, E. coli transformed with a pAED4 plasmid lacking the cspBPi insert was used. One ml aliquot of liquid culture was placed at −20°C for 2h. The frozen cells were taken out of the freezer and put on ice for 1h to be thawed. This process was performed in duplicates and repeated up to three cycles. Aliquots were taken at each cycle of freeze-and-thaw, and colony-forming units (CFU) were counted after incubation on LB plates at 37°C for 24h. The data were collected from five independent experiments and shown as an average for each point.
Purification of CspBPiCspBPi were overexpressed at 37°C in E. coli BL21(DE3) as described above, except that the cultures were scaled-up to 1L liquid LB media. Cells were harvested by centrifugation, and resuspended in 40ml of 10mM phosphate buffer, pH 6.5. Cells were lyzed by sonication using a Bandelin Sonoplus HD2200 ultrasonic homogenizer (Berlin, Germany) as described for CspAPi.16 The supernatant fraction was loaded on a Q-sepharose™ (Amersham Bioscience Co.) fast flow ion exchange column equilibrated with the same buffer. CspBPi protein was eluted with a 0–0.5M NaCl gradient. Concentrations of proteins were determined using Bio-Rad DC (detergent compatible) protein assay kit, and the purity of proteins was analyzed by 20% SDS-PAGE.
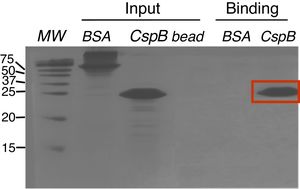
Oligo(dT)-cellulose binding assaysPurified CspBPi protein was dialyzed against binding buffer (10mM Tris–HCl, pH 8.0, 1mM EDTA, 50mM KCl, and 7.4% glycerol). Fifty μl of oligo(dT)-cellulose type 7 beads (Amersham Bioscience Co.) were incubated with 50μg of CspBPi protein at 4°C for 4h. The resins and bound proteins were collected by brief centrifugation, and washed twice with the binding buffer. In parallel, 50μg of bovine serum albumin (BSA) protein, instead of CspBPi protein, was used as the experimental control. Co-precipitated proteins were analyzed by 20% SDS-PAGE and Coomassie brilliant blue R250 staining.
Rescue of cold-sensitive E. coli BX04 strainE. coli quadruple csp deletion strain BX04 cells harboring pAED,18 pAED-cspAPi,16 pAED-cspBPi, or pAED-cspCPi16 were grown in the liquid culture to an OD600 of 0.4. The cells were then streaked onto LB plates containing 0.1mM IPTG, and incubated at temperatures ranging from 16 to 37°C. After 15h at 37°C, 24h at 25°C, or 60h at 16°C, the growth of BX04 cells on the plates was observed.
ResultsCloning and sequence analysis of the cspB gene from P. irgensii KOPRI 22228P. irgensii KOPRI 22228 was isolated from Arctic Sea sediments and grown as previously reported.16 This bacterium is a psychrophile, with an optimum growth temperature of 10°C. In an effort to elucidate the roles played by Csps in psychrophilic bacteria, DNA fragments from this bacterium, which contain a region encoding a conserved CSD, were cloned. A new csp-homologous gene 0.45kb in size was obtained and named cspBPi.
A BLAST nucleotide homology search of cspBPi was performed: cspBPi exhibited low homology to the canonical cspAEc (15.8%) and E. coli cspD (12.4%). The deduced amino acid sequence of cspBPi encoded a protein 150 residues in length, and the CspBPi sequence was aligned to previously studied Csp sequences from polar bacteria: CspA from Streptomyces sp. AA8321 (CspASt),20 CspA from Psychromonas arctica KOPRI 22215 (CspAPa)21 and CspAPi and CspCPi from P. irgensii KOPRI 22228.16 All Csps have a typical β-barrel CSD composed of five β-strands, but CspBPi has a unique extra domain at its N-terminal end (Fig. 1). CSD is highly conserved among many bacterial Csps, and the three-dimensional structures of some Csps, including CspAEc and Bacillus subtilis CspB, have been reported.22,23 CSD is believed to mediate nucleic acid binding. In particular, two RNA-binding motifs, RNP1 (with consensus sequence K-G-F-G-F-I) and RNP2 (with consensus sequence V-F-V-H-F) are crucial for binding to RNA or ssDNA (boxed in Fig. 1).24 In RNP2 of CspBPi, the first and the second Val residues were replaced with Tyr and Thr, respectively, and the last Phe residue was replaced with Val. However, all three Phe residues (Phe-15, Phe-17, and Phe-28; residue numbers according to the canonical CspAEc) in the RNA-binding motifs, which are considered to be necessary for nucleic acid-binding activity,24,25 were conserved in CspBPi. Sequence analysis of CspBPi suggested that the protein may bind to RNA or ssDNA through a canonical cold shock domain β-barrel structure. Meanwhile, the additional N-terminal domain of this protein did not show any noticeable homology to other proteins.
Sequence alignment of various Csp proteins from polar organisms. Abbreviations for bacterial Csps whose amino acid sequences were analyzed here: S. sp. CspA, Streptomyces sp. AA8321 CspASt20; P. irgensii CspA and CspC, Polaribacter irgensii KOPRI 22228 CspAPi and CspCPi, respectively16; P. irgensii CspB, P. irgensii KOPRI 22228 CspBPi from this study; Ps. arctica CspA, Psychromonas arctica KOPRI 22215 CspAPa.21 The amino acid sequences of Csp proteins were aligned using the default settings of CLUSTAL W.27 Below the protein sequences is a key denoting conserved sequence (*), conservative mutations (:), semi-conservative mutations (.), and non-conservative mutations (). Gaps indicated by hyphens (-) were introduced to improve alignment. The RNA-binding motifs RNP1 and RNP2 are boxed.
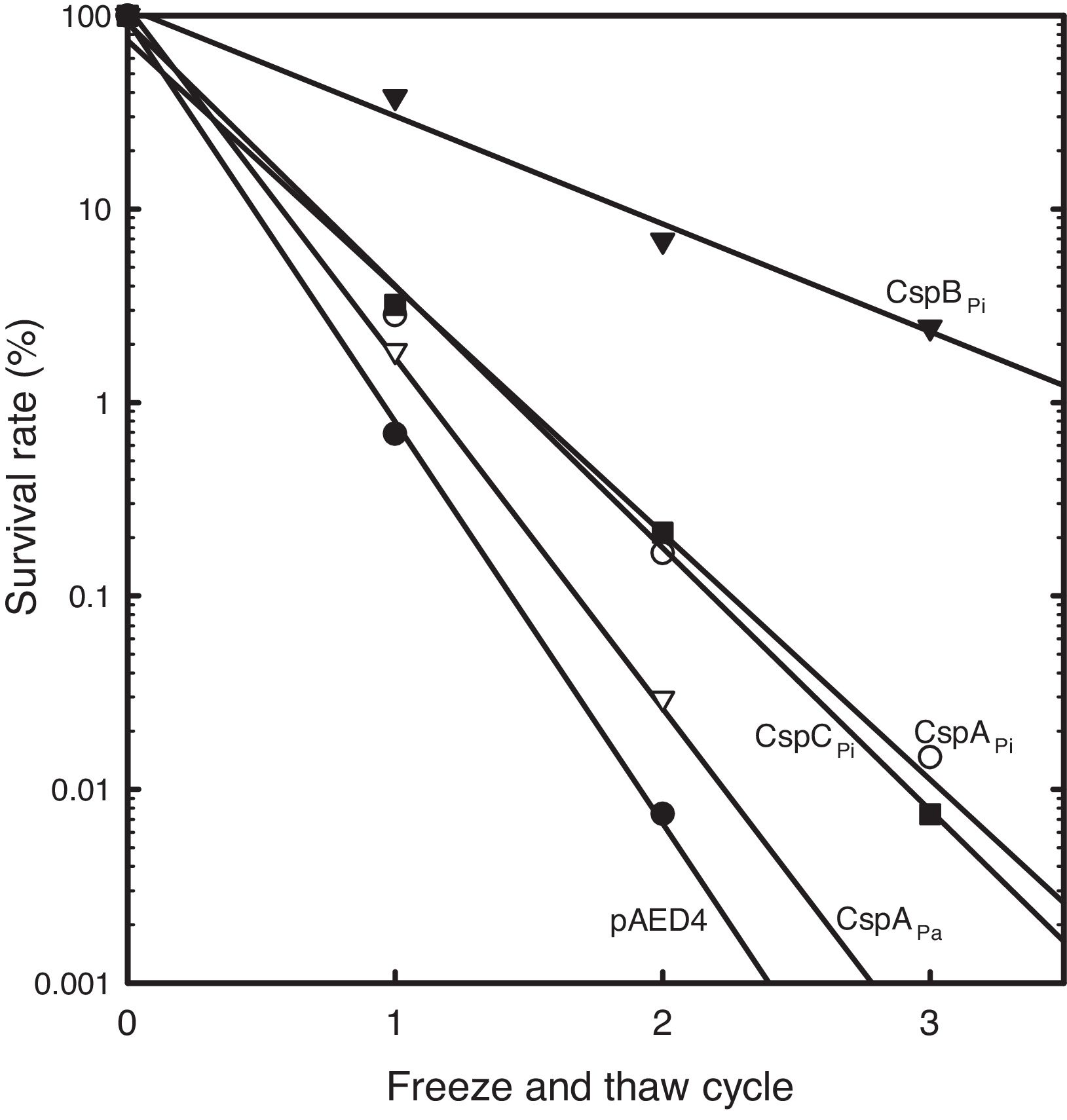
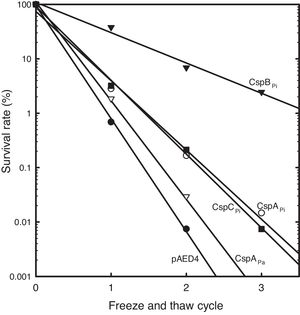
To study the roles of Csps from psychrophilic bacteria, cold resistance of the hosts harboring csp genes, was examined. Csp-overexpression was induced by the addition of IPTG to mid-log phase liquid cultures of cells carrying a csp-expression vector. When E. coli cells harboring pAED4 were frozen and thawed once, less than 1% of the original cells survived. Following repeated cycles of freezing and thawing, the number of surviving cells decreased almost exponentially. Overexpression of previously reported csp genes from polar bacteria increased freeze-survival rates of the hosts only moderately: the Ps. arctica CspAPa-expressing cells exhibited a slightly increased survival rate and the CspAPi or CspCPi-overexpressing cells showed more than five-fold increase in the survival rates in the first freeze-thaw cycle (Fig. 2).16,21 Surprisingly, the CspBPi-overexpressing cells showed an extraordinary increase in freeze-tolerance: more than fifty fold the number of cells survived the first freeze–thaw cycle and the number of surviving cells increased to greater than 100000-fold after three cycles of freezing and thawing compared to the number of surviving pAED4-carrying cells (Fig. 2).
Greatly increased cold-resistance of CspBPi-overexpressing cells. The survival rates of various Csp-overexpressing cells following cycles of freeze-and-thaw are shown. The number of viable cells prior to freezing was set at 100%. ●, Control pAED4-carrying cells; ▿, CspAPa-overexpressing cells; ○, CspAPi-overexpressing cells; ■, CspCPi-overexpressing cells; ▾, CspBPi-overexpressing cells.
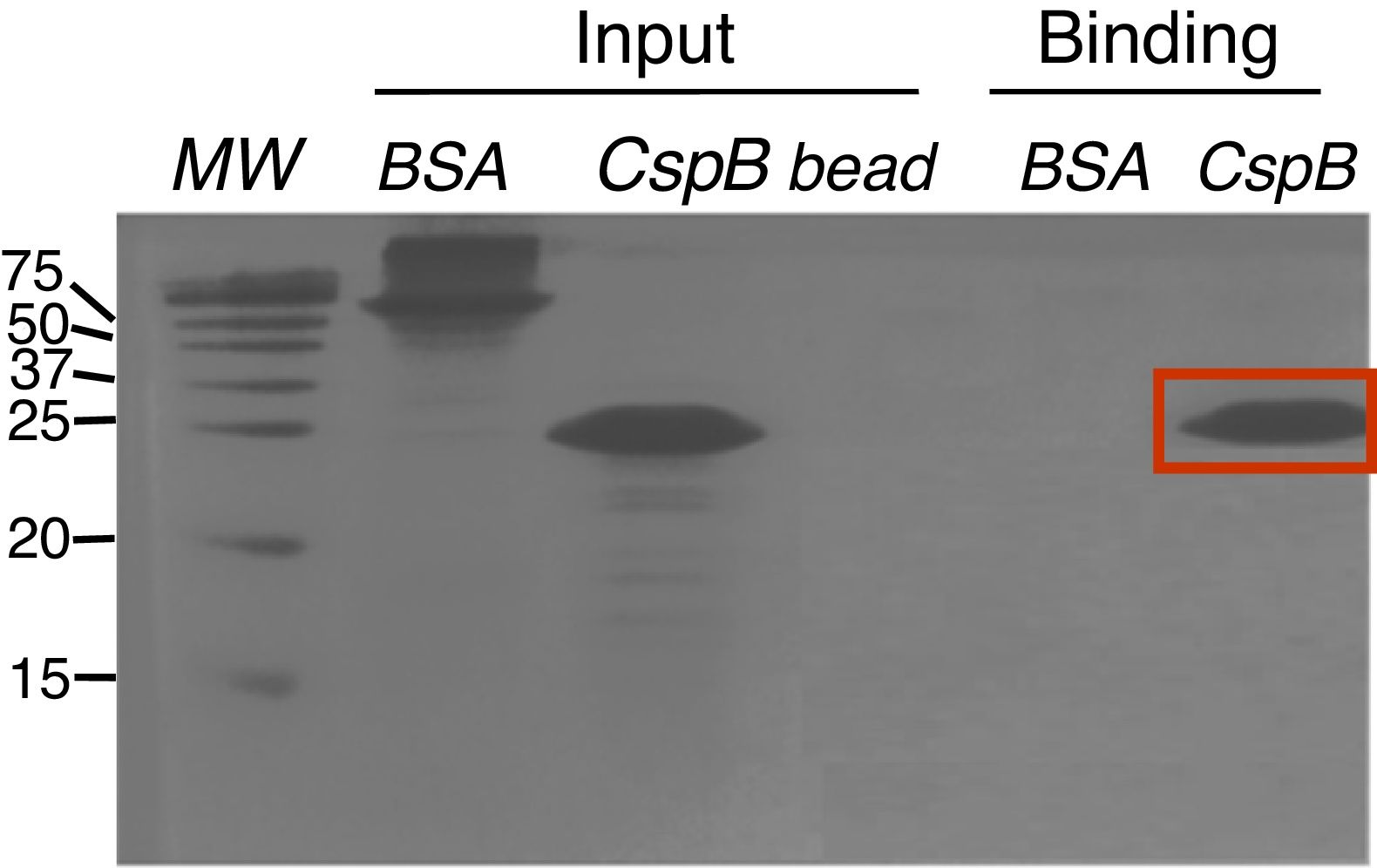
Since CspBPi contains CSD and the conserved Phe residues in the RNP1 and RNP2 sequence motifs, which suggest single-stranded RNA or DNA binding activity, the functionality of the single-stranded nucleic acid-binding motifs of this protein was examined. Upon incubation of E. coli BL21 cells harboring pAED-cspBPi with 0.1mM IPTG, a protein band with an apparent molecular mass of 23kDa on SDS-PAGE was observed. CspBPi was expressed in soluble form and purified by anion-exchange column chromatography. Partially purified CspBPi was incubated with oligo(dT)-cellulose beads at 4°C for 4h and subjected to a brief centrifugation. When the reaction products were analyzed by 20% SDS-PAGE, CspBPi was bound to the oligo(dT)-cellulose and co-precipitated with the resins, while neither bovine serum albumin used at the same concentration nor contaminating proteins from the CspBPi preparation were co-precipitated (Fig. 3). The result suggests that CspBPi binds ssDNA.
Oligo(dT)-binding activity of CspBPi. CspBPi was incubated with oligo(dT)-cellulose at 4°C for 4h. Proteins bound to beads were collected by centrifugation. Lanes: MW, precision plus protein standards (Bio-Rad Laboratories Inc.; size of each protein band is shown in kDa at left of the gel); BSA, bovine serum albumin; CspB, CspBPi; bead, oligo(dT)-cellulose only. The migration position of the bound CspBPi protein is indicated with a red box.
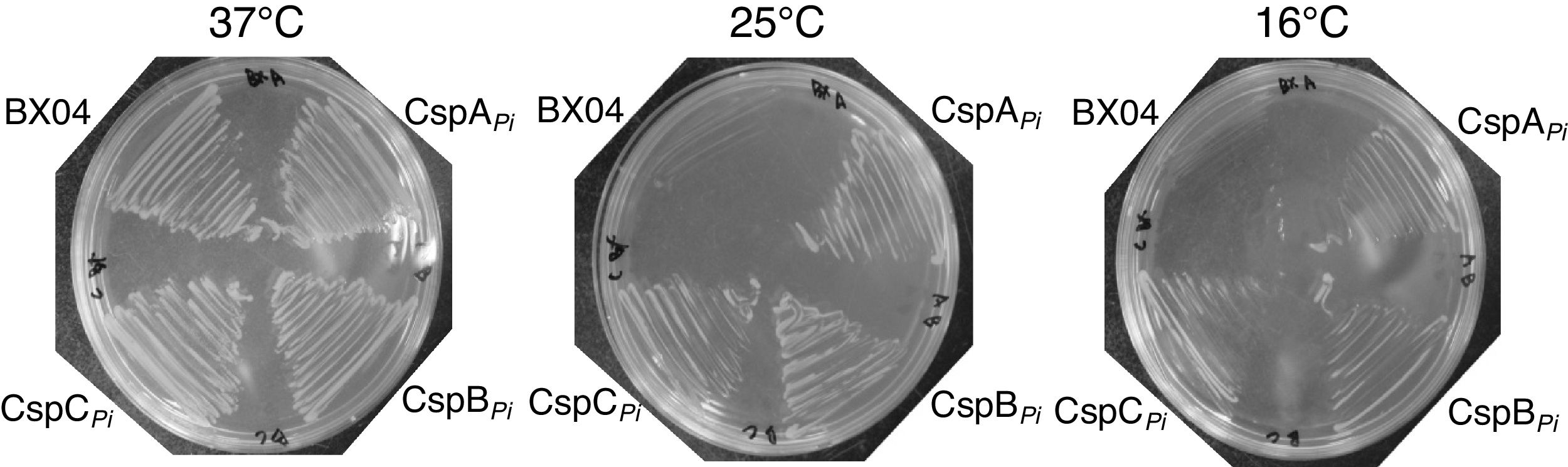
Since overexpression of CspBPi greatly increased the cold resistance of wild-type E. coli (Fig. 2), the ability of CspBPi to suppress the cold-sensitivity of the E. coli quadruple csp deletion strain BX04 (ΔcspA, ΔcspB, ΔcspG, and ΔcspE) was also examined. Mid-log phase cultures of BX04 cells harboring pAED4, pAED-cspAPi, pAED-cspBPi, or pAED-cspCPi were streaked onto LB plates containing 0.1mM IPTG and incubated at temperatures ranging from 16 to 37°C. As described previously,17 the growth of BX04 cells was comparable to other clones at 37°C, but the growth was extremely retarded at 16°C (Fig. 4). Meanwhile, overexpression of any Csps from P. irgensii complemented the cold-sensitivity of BX04 at 25°C and at 16°C (Fig. 4). When the growth of BX04 was followed by OD600, overexpression of CspBPi promoted the growth at low temperatures at slightly higher levels than CspAPi or CspCPi did (data not shown).
Overexpression of CspBPi rescued cold-sensitive phenotype of csp quadruple-deletion E. coli strain, BX04. BX04 cells harboring pAED, pAED-cspAPi, pAED-cspBPi, or pAED-cspCPi were grown in the liquid culture to an OD600 of 0.4. The cells were then streaked on LB plates containing 0.1mM IPTG, and incubated at temperatures ranging from 16 to 37°C.
Cold temperatures affect all physical-chemical parameters of living organisms, and effects include decreased solute diffusion rates, ice crystal formation, dehydration, decreased membrane fluidity, slow enzyme reaction rates, stable secondary structure formation in nucleic acids, and cold-denaturation of proteins. Psychrophilic organisms from cold ecosystems have evolved biological means to circumvent these challenges. Although transcription and translation of most genes are nearly stopped upon sudden temperature drops in mesophiles, expression of cold-shock genes are selectively induced.1 Meanwhile, corresponding genes in psychrophiles are more consistently expressed, instead of being transiently induced during the cold acclimation phase,3 suggesting that Csps play important roles for survival in cold environments.
In an effort to understand the mechanisms played by psychrophiles to adapt to cold biosphere, this study focused on the functions of Csps from polar bacteria. Although dozens of csp genes from psychrophiles have been identified by genomic/metagenomics approaches, their roles on cold-adaptation were elucidated only in very limited cases. Heterogeneous expression of Csps from polar microorganisms resulted in various effects on cold adaption of their recombinant hosts. Certain Csps failed to increase cold-resistance of their hosts: CspASt from an Antarctic Streptomyces neither increased the cold-resistance of wild-type E. coli, nor that of the E. coli quadruple csp deletion strain BX04.20 Instead overproduction of CspASt inhibited DNA replication, as non-canonical E. coli CspD does, suggesting a role for CspASt in halting DNA replication until the cell adjusts itself upon sudden temperature drops.20 Similarly, overexpression of a Csp from an Antarctic haloarchaeon Halorubrum lacusprofundi did not suppressed the cold sensitivity of BX04.26 Meanwhile, it has been reported that the overexpression of CspAPa increased the cold-resistance of wild-type E. coli, but not of the E. coli quadruple csp deletion strain BX04.21 Therefore, the contribution of CspAPa to the cold survival of their recombinant hosts seemed relatively modest. Other Csps from psychrophilic organisms were more effective in increasing cold tolerance of their hosts; the overexpression of CspAPi or CspCPi not only increased the cold-survival rates of wild-type E. coli by more than five-fold following one cycle of freezing and thawing, but also rescued cold-sensitive phenotype of BX04.16 Overexpression of Csps from a stenopsychrophilic archaeon Methanogenium frigidum or a deep sea planktonic archaeon Crenarchaeota also complemented cold susceptibility of E. coli BX04.26 Since no quantitative data on freeze-tolerance conferred by these proteins were provided, more detailed studies would be necessary to compare their effect with that of CspBPi. However, overexpression of any E. coli Csps, except CspDEc, also rescued cold-sensitive E. coli BX04,17 and overexpression of CspAEc increased survival rates of the wild-type E. coli upon freezing and thawing at a comparable level to CspAPi or CspCPi (data not shown). These results show that the ability to confer cold resistance to their hosts is not the unique characteristics of Csps from psychrophiles: previous studies rather suggest that several Csps from psychrophiles have retained sufficient similarity throughout evolution to be able to function effectively in mesophiles to confer cold tolerance.
Surprisingly, overexpression of CspBPi greatly increased the cold tolerance of its recombinant host to an unprecedented level (Fig. 2). Overexpression of CspBPi not only induced a more than fifty fold increase in freeze-tolerance in wild-type E. coli (Fig. 2), but also noticeably promoted the growth of the E. coli quadruple csp deletion strain at low temperatures (Fig. 4). Elucidating the detailed mechanisms involved in freeze-tolerance conferred by CspBPi would be of great academic interest and will be pursued in a future study. CspBPi possesses cold shock domains at the C-terminal region (Fig. 1) and shares the basic characteristics of Csps, including the ability to bind single-stranded nucleic acids, as indicated by oligo(dT)-binding assays (Fig. 3). One possibility is that the CSD of CspBPi may have greatly improved activity, for example as a RNA chaperone, maintaining single stranded nucleic acid structures at extremely low temperatures and allowing efficient transcription and translation. On the other hand, CspBPi has an extra domain on its N-terminal region, and it is also possible that this unique region plays a distinct role in conferring an extraordinary increase in cold survival ability. Sequence homology search has identified several cspBPi-homologous genes, encoding both N-terminal extra domain and CSD, from evolutionarily related marine Flavobacteriaceae, including Polaribacter sp. MED152, Lacinutrix sp. 5H-3-7-4, Winogradskyella sp. PG-2, and Maribacter sp. HTCC2170. However, their functional roles on cold tolerance or transcriptional/translational regulation upon temperature downshifts have not been studied yet. It will be interesting to test freeze-tolerance of hosts overexpressing these dual domain Csp-like proteins.
Introduction of CspBPi into other organisms has potential industrial applications, including increased cold-resistance of nitrogen-fixing bacteria, such as Rhizobium and cyanobacteria, and the survival of starter cultures after storage at freezing temperatures. The long-pursued development of frozen dough may be achievable by improving the freeze-tolerance of baker's yeast. The possible application of CspBPi does not need to be limited to industrially important microorganisms and it may be introduced into plants to enhance their viability at low temperatures and increase their economic value.
Conflicts of interestThe authors declare no conflicts of interest.
The authors greatly appreciate the gift of E. coli BX04 from Dr. Sangita Phadtare and Dr. Massayori Inouye (UMDNJ). This work was supported by the Korea Research Foundation Grant funded by the Korean Government (KRF-2014-011146 and KRF-2015R1D1A1 A01058206).