The virulence genes in invasive aspergillosis (IA) have not been analyzed adequately. The present study was designed to evaluate the expression of gpaB and sidA genes, which are important virulence genes in Aspergillus spp. from bronchoalveolar lavage (BAL) samples. Direct examination and culture on Czapek Agar and Sabouraud Dextrose Agar media were performed for 600 BAL specimens isolated from patients with possible aspergillosis. A Galactomannan ELISA assay was also carried out. The expression levels of the gpaB and sidA genes in isolates were analyzed using quantitative real-time PCR (qRT-PCR). We identified 2 species, including Aspergillus flavus (A. flavus) and Aspergillus fumigatus (A. fumigatus) in 25 positive samples for invasive aspergillosis as validated using GM-ELISA. A. flavus is the main pathogen threatening transplant recipients and cancer patients worldwide. In this study, A. flavus had low levels of the gpaB gene expression compared to A. fumigatus (p=0.006). The highest sidA expression was detected in transplant recipients (p=0.05). There was no significant correlation between sidA expression and underlying disease (p=0.15). The sidA and gpaB gene expression patterns may provide evidence that these virulence genes play important roles in the pathogenicity of Aspergillus isolates; however, there are several regulatory genes responsible for the unexpressed sidA and gpaB genes in the isolates.
Invasive aspergillosis (IA) is a severe disease and one of the primary causes of mortality among immunocompromised individuals, including cystic fibrosis and cancer patients, as well as transplant recipients.1 A variety of Aspergillus species act as causative agents of IA, but A. fumigatus and A. flavus are the major causes of the disease. In fact, the small size of their conidia facilitates the entry of fungi into the alveolar cavity.2–4 If we want to map the gene network, which is associated with the progression of IA in immunocompromised individuals, we will need detailed information related to the virulence gene profiles of Aspergillus spp.
Several putative virulence factors contributing to the pathogenesis of aspergillosis,5 have been identified, including pigment production, adhesion molecules on the cell surface, hydrolytic enzymes and toxin secretion. Additionally, it has been proven that a network of genes is required to support the virulence of the Aspergillus spp.6
G-proteins, involved in signal transduction pathways, and siderophores are encoded by specific genes in the Aspergillus genome7 and they are essential for A. fumigatus to reprogram its physiology in response to the environment and iron uptake.8,9 G protein signaling and the way by which it is regulated have been studied in a number of models in the past decade. Studies on the pathogenic fungi have opened a new avenue to the concept of the regulatory mechanisms of development, virulence/pathogenicity establishment, mating, and generation of secondary metabolites in different fungi. Certainly, A. fumigatus Gβ and Gγ subunits are inevitable for viability and vegetative growth, because gene deleted mutants are extremely defective in germination and vegetative growth.10 gpaB genes encode G-proteins that have a central role in the integrity, morphogenesis, and virulence of A. fumigatus, as well as eliciting the host's immune response. A previous study demonstrated that the gpaB gene mutant strain of Aspergillus is avirulent in animal-infected IA models.11
In a study performed by Gehrke et al. it was shown that deletion of two putative G-protein-coupled receptors, GprC and GprD, caused devastating growth defects. Furthermore, hyphal extension and germination were decreased. They showed that the mutants strain displayed an attenuation of virulence in a murine infection model.12
Askew and colleagues showed the gene networks with probable roles in the integrity of the cell wall and environmental change-related signaling pathways.13 Siderophore proteins are encoded by sidA genes and play important roles in several vital processes, including germination, oxidative stress resistance, and iron storage in Aspergillus spp.9,14 Interestingly, it has been shown that A. fumigatus exploits sidA gene expression for virulence and pathogenicity.15,16
Hissen indicated that a rescued strain of Aspergillus (Delta sidA+sidA) exhibited siderophores and had similar growth to the wild type on Iron-limited media. In contrast to the wild-type and other rescued strains, it was reported that the Delta sidA strain was virulent in a mouse model of IA. It demonstrates that sidA plays a pivotal role in the pathogenesis of A. fumigatus.15 Previous studies have already described the role of genes in the pathogenesis of Aspergillus. Not only it is not well-defined which gene is exactly involved in Aspergillus pathogenesis but also the exact function of each gene has remained unknown.6 The expression of sidA and gpaB genes in Aspergillus spp. was not completely defined in BAL specimens isolated from immunosuppressive subjects. Therefore, regarding the importance of gpaB and sidA genes in virulence of Aspergillus spp. either in vitro or in vivo. This study focused on gpaB and sidA gene expressions in A. fumigatus and A. flavus for the first time, as the primary genes involved (incriminated) in the pathogenesis of Aspergillus spp. in BAL samples isolated from immunocompromised and hospitalized subjects predisposed to developing IA.
Materials and methodsPatient characteristicsBronchoalveolar lavage (BAL) samples were collected by a pulmonologist via bronchoscopy from 600 patients with underlying disease and susceptible to IA between 2014 and 2015 at Masih Daneshvari Hospital, Tehran, Iran. The patient populations include cancer patients (leukemia, liver), transplant recipients (bone marrow, lung, and kidney), chronic obstructive pulmonary disease (COPD), hemoptysis (with an immunosuppressive condition), and allergic bronchopulmonary aspergillosis (ABPA) patients taking corticosteroid.17 All of the participants filled informed consent. The patients’ medical records and other data, including age, gender, and type of disease were collected and recorded in a database. These data were kept fully anonymous. The study protocol was approved by the Research Ethics Committee of Iran University of Medical Sciences (IUMS). All BAL samples were collected twice per patient.
Specimen cultureThe BAL samples were immediately transferred to a medical mycology laboratory. The specimens were used in a direct smear with KOH 10% and then cultured on Czapek Agar and Sabouraud Dextrose Agar media (2% glucose, 1% peptone, 0.05% chloramphenicol, 2% agar) (Merck, Germany) for 7 days at 25°C. The growth rate of Aspergillus was checked and the species were identified using conventional phenotyping methods such as colony colors and slide cultures.
Confirmation of aspergillosis using GM-ELISAThe GM-ELISA (Platelia Aspergillus; Bio-Rad, Edmonds, WA) was performed on positive BAL samples, according to the manufacturer's recommendations.18 Briefly, 100μL of the Platelia treatment solution was added to 300μL of the BAL specimen and heated at 100°C for 3min before undergoing centrifugation. The supernatant and the horseradish peroxidase-labeled monoclonal antibody (EBA-2) were incubated at 37°C in antibody-precoated microplates. The plates were washed and then incubated with a substrate chromogen reaction solution. The reaction was stopped by the use of 100μL of sulfuric acid and after 30min, the optical density (OD value) of each well was read using a microplate reader at 450nm. One well for the negative control, two wells for the cut-off control, and one well for the positive control were used. The results of Galactomannan ELISA assay are expressed as “galactomannan index” (GMI=OD sample/mean OD cut-off control), by comparison to the “cutoff” control. A GMI index of ≥1.0 in two consecutive samples was considered positive.
Culture of Aspergillus spp.For RNA extraction, the Aspergillus spp. was cultured in a Yeast Sucrose Broth (YSB) medium (Merck, Germany) at 25°C for 48–72h. The optimal growth rate was determined based on the expansion of mycelium on the surface of the medium.
RNA extraction, cDNA synthesis, and quantitative real-time PCRRNA contents within all the samples were extracted using the RNA-X PLUS solution (Cinaclon, cat=RN7713C-s). After disrupting the Aspergillus spp. cell wall using liquid nitrogen, the RNA-X PLUS solution was added to the sample. Chloroform was added after centrifugation of liquid at 2000rpm for 5min, the aqueous phase was transferred to a new tube. The equal volume of isopropanol added, then the tube was centrifuged at 2000rpm for 5min and washed with 75% ethanol, the semidried pellet of RNA was dissolved in DEPC treated water. The RNA was treated with DNase (DNase I) (Invitrogen, Cat No 18068015) to remove genomic DNA contamination. The quality and quantity of the extracted RNA, before and after treatment, were examined using a gel electrophoresis and BioPhotometer plus (Germany), respectively.
First-strand cDNA was synthesized using a high-capacity cDNA reverse transcription kit (Hyper script reverse transcriptase, Gene All, Cat No 601-100), according to the manufacturer's protocol. The cDNA samples were kept at −80°C until being used. The expression of the gpaB and sidA genes was quantified by real-time PCR analysis, as previously described.19 The reactions contained 2μL of template cDNA, 0.6μL of each specific primer (10pmol) for the sidA and gpaB genes, 10μL of SYBR Green master mix, and 6.8μL of DEPC water in a final volume of 20μL. The results were normalized using β-tubulin amplifications run on the same plate as the house keeping gene. The real-time PCR system was programmed to an initial denaturation and polymerase activation for 2min at 95°C, denaturation for 15s at 95°C, and annealing and elongation for 1min at 62°C over 35 cycles using a Corbet thermo cycle (RG-6000, Australia). Specific primers for quantitative real-time RT-PCR were designed with online primer design software (Genscript, Piscataway, NJ). For determination of primers efficiency values, we papered the serial dilutions (25ng, 5ng, 1ng, 0.2ng, 0.04ng per microliter) of the positive control for sidA and gapB genes. Then, the primers efficiency values were determined using a Corbet thermo cycle (RG-6000, Australia) (Table 1). The specificity of the primers was assessed with genomic DNA from different Aspergillus species. The standard species of A. fumigatus and A. flavus (as control group) were used in relative quantitative method using real-time PCR. The level of sidA and gapB expressions was divided into either down-regulation (≤mean of gene expression) or up-regulation (>mean of gene expression), as described previously.20
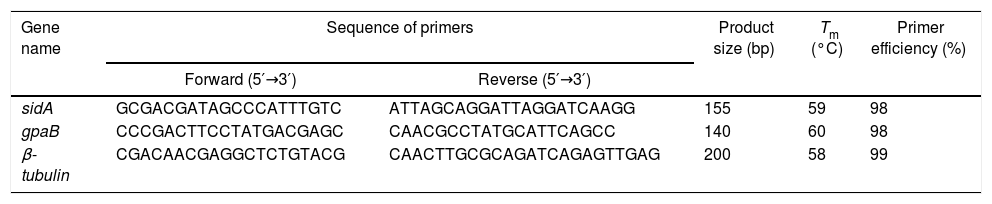
Sequence of primers in quantitative real-time PCR.
| Gene name | Sequence of primers | Product size (bp) | Tm (°C) | Primer efficiency (%) | |
|---|---|---|---|---|---|
| Forward (5′→3′) | Reverse (5′→3′) | ||||
| sidA | GCGACGATAGCCCATTTGTC | ATTAGCAGGATTAGGATCAAGG | 155 | 59 | 98 |
| gpaB | CCCGACTTCCTATGACGAGC | CAACGCCTATGCATTCAGCC | 140 | 60 | 98 |
| β-tubulin | CGACAACGAGGCTCTGTACG | CAACTTGCGCAGATCAGAGTTGAG | 200 | 58 | 99 |
The mean of gene expression was used as the cut-off for categorization in low and high expression groups. Relative expression was expected using the formula 2−ΔΔCt. The expression of the sidA and gpaB genes was normalized to β-tubulin expression. The data were analyzed using SPSS software, version 20 (SPSS, Chicago, IL, USA). The association between the level of gpaB and sidA expressions and patient characteristics were explored using either Pearson's chi-square or Fisher's exact test. Variations in the expression of sidA and gpaB between immunosuppressive diseases were assessed using the Mann–Whitney U test. A p-value of ≤0.05 was considered to be statistically significant.
ResultsPopulation studyTwenty-five positive cases were detected through the screening of 600 BAL samples received from studied patients. The A. flavus isolates were found in 17 (68%) of the cases, and A. fumigatus isolates were also detected in 8 (32%) of the cases. Twelve (48%) patients were male, and 13 (52%) were female (M/F ratio=0.92); they ranged in age from 40 to 83 years (mean=59 and median=60 years). The obtained results from patient's data showed that 9 (36%) of the patients had COPD, while transplant recipients and cancer patients were found in similar frequencies (24%). In addition, hemoptysis and ABPA were detected in only 2 cases (8%) (Table 2). The results of GM-ELISA test indicated that GM antigen was successfully detected in the serum of the patients with positive Aspergillus spp. in BAL samples, the result analyzed according to GM index.
Patients’ characteristics and correlation between the levels of sidA and gpaB gene expressions (bold values are significant).
| Patient data | Total no. | sidA expression (mean=2.4) | p-Value | gpaB expression (mean=33) | p-Value | ||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
| Age (mean=59) | |||||||
| ≤59 | 14 (56) | 10 (71) | 4 (29) | 0.54 | 13 (93) | 1 (7) | 0.17 |
| >59 | 11 (44) | 9 (82) | 2 (18) | 8 (73) | 3 (27) | ||
| Gender | |||||||
| Male | 12 (48) | 9 (75) | 3 (25) | 0.9 | 10 (83) | 2 (17) | 0.93 |
| Female | 13 (52) | 10 (77) | 3 (23) | 11 (85) | 2 (15) | ||
| Disease | |||||||
| Transplant recipient | 6 (24) | 3 (50) | 3 (50) | 0.15 | 6 (100) | 0 (0) | 0.07 |
| Cancer | 6 (24) | 6 (100) | 0 (0) | 6 (100) | 0 (0) | ||
| COPDa | 9 (36) | 8 (89) | 1 (14) | 6 (67) | 3 (33) | ||
| Hemoptysis | 2 (8) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | ||
| ABPAb | 2 (8) | 1 (50) | 1 (50) | 2 (100) | 0 (0) | ||
| Aspergillus spp. | |||||||
| flavus | 17 (68) | 12 (71) | 5 (29) | 0.28 | 17 (100) | 0 (0) | 0.006 |
| fumigatus | 8 (32) | 7 (88) | 1 (12) | 4 (50) | 4 (50) | ||
For analysis of gpaB gene expression, firstly, the expression level of gpaB gene in each case was normalized with β-tubulin, as housekeeping gene and the final gene expression level has been calculated (2−ΔΔCt). Then, mean value of gpaB gene expression was selected as a cut-off value to categorize the level of gene expression. The mean gpaB expression (mean=33) was chosen as a cut-off value to classify the samples into two categories: each case with lower than of mean gene level was considered as the down-regulation expression, and each case with higher than of mean gene level was considered as the up-regulation expression. Down-regulation gpaB expression was found mainly in cancer patients and transplant recipients; these same patients showed no instances of up-regulation gpaB expression (Table 2).
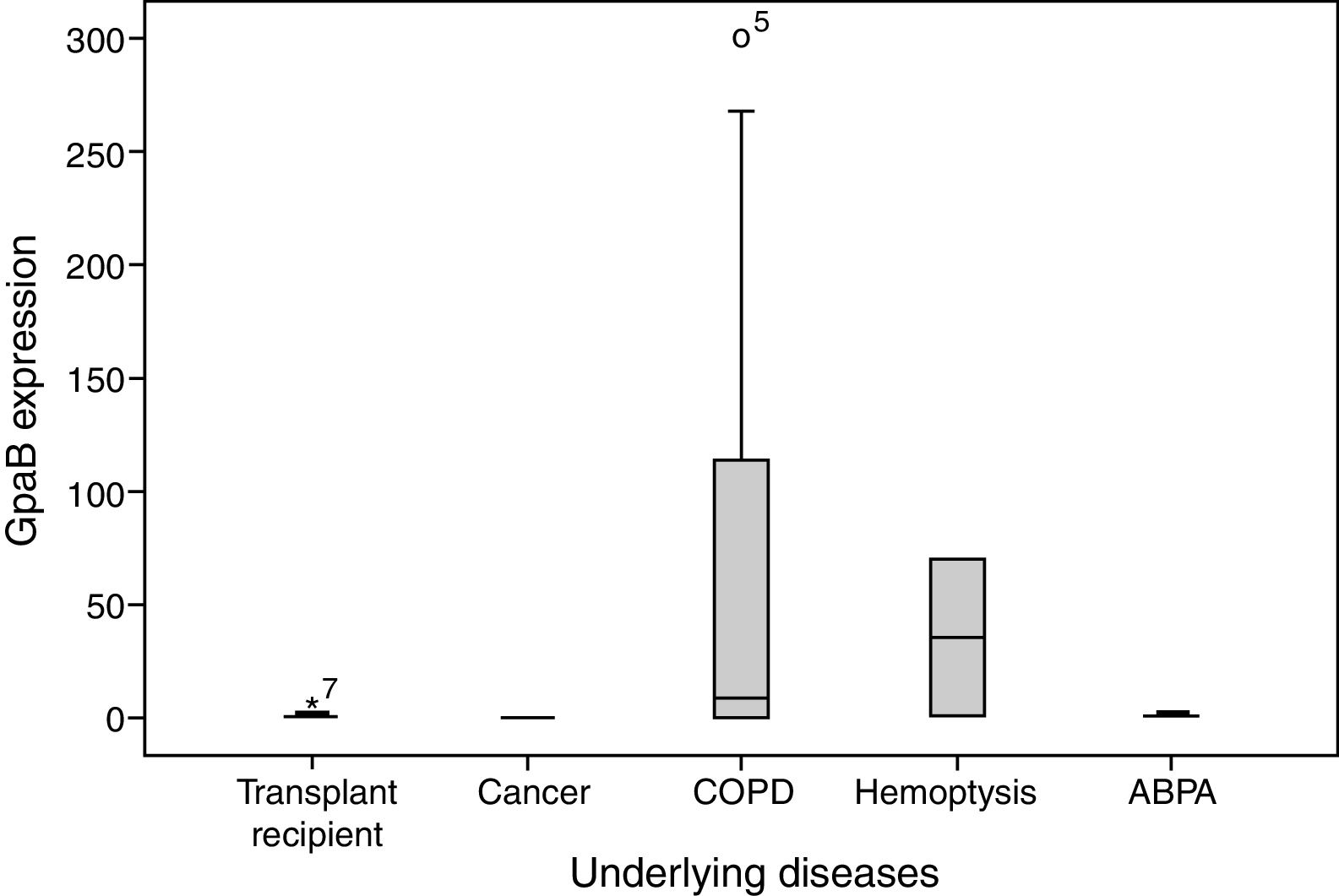
Correlations between the level of gpaB expression and patient characteristics were explored using either Pearson's chi-square or Fisher's exact test. A significant association was observed between gpaB expression and Aspergillus spp. using fisher exact test, indicating that A. flavus expressed lower levels of gpaB than A. fumigatus (p=0.006, Table 2). Moreover, a trend became evident between decreased gpaB expression and underlying disease using Chi-square test (p=0.07, Table 2). No correlation was found between gpaB expression and other patient characteristics, including age and gender (p=0.17 and p=0.93, respectively). Among cases, a comparison of the differences between expression using the Mann–Whitney U test demonstrated a marginal trend between cancer patients and both hemoptysis patients and transplant recipients (p=0.08 and p=0.09, respectively). In addition, a marginal trend was found between transplant recipients and hemoptysis patients. (p=0.08) (Fig. 1).
Analysis of gpaB expression levels in patients using Mann–Whitney U test. On the basis of the standard definitions, Box-plot shows median (bold line), interquartile line (box), outliers (circle), and extreme observations (star). COPD, chronic obstructive pulmonary disease; ABPA, allergic bronco pulmonary aspergillosis.
For analysis of sidA gene expression, firstly, the expression level of sidA gene in each case was normalized with β-tubulin, as housekeeping gene and the final gene expression level has been calculated (2−ΔΔCt). Then, mean value of sidA gene expression was selected as a cut-off value to categorize the level of gene expression. The mean of sidA expression (mean=2.4) was chosen as a cut-off value to classify the samples into two categories: Each case with lower than of mean gene level was considered as the down-regulation expression, and each case with higher than of mean gene level was considered as the up-regulation expression. Low sidA expression was frequently observed in COPD and cancer cases (Table 2). Correlations between the level of sidA expression and patient characteristics were explored using either Pearson's chi-square or Fisher's exact test. Down-regulation of sidA expression was mainly found in A. flavus, but no significant association was found between sidA expression and Aspergillus spp. (p=0.28, Table 2). Although down-regulation of sidA expression was detected in cancer and COPD patients, no significant correlation was found between sidA expression and underlying disease (p=0.15). Among the patients, a comparison of the differences between sidA expression using the Mann–Whitney U test demonstrated a significant difference between cancer patients and transplant recipients (p=0.05), and a marginal trend between cancer patients and both hemoptysis and ABPA patients (p=0.08 and p=0.08, respectively) (Table 2 and Fig. 2).
Analysis of sidA expression levels in patients using Mann–Whitney U test. On the basis of the standard definitions, Box-plot shows median (bold line), interquartile line (box), outliers (circle), and extreme observations (star). COPD, chronic obstructive pulmonary disease; ABPA, allergic bronco pulmonary aspergillosis.
A. flavus was the main pathogen in cancer patients (100%) and transplant recipients (83%). A. flavus and A. fumigatus play similar roles in other cases, including COPD, hemoptysis, and ABPA.
DiscussionThe incidence of IA has increased in recent years in patients with predisposing factors.21A. fumigatus and A. flavus are known as important agents that can cause IA with enhanced morbidity and mortality in immunocompromised patients, such as chronic obstructive pulmonary disease (COPD) and cancer patients, patients taking corticosteroids and antimicrobial prophylactic drugs, and transplant recipients.22,23 Due to the suppressed immune systems of such individuals, the Aspergillus conidia can pass through the alveolar epithelial cells by conidiation and germination, thereby inducing pathogenicity in patients.24,25 Furthermore, the expression of some Aspergillus genes has been observed to be changed with extensive prophylaxis with fluconazole, which increases the virulence potency.26
The functions and character of the putative virulence factors of A. fumigatus are still under investigation. Although several genes have been discovered, there are a lot of other genes which are responsible for the regulation of virulence gene expression. It often makes it impossible to designate with certainty which of them contribute to the mechanism of pathogenesis.12 Regarding the dominant role of virulence genes in the pathogenesis of IA, there is no comprehensive information concerning gene expression in Aspergillus, although a few studies have investigated this issue in recent years.
Our findings demonstrated the up-regulation expression of the gpaB gene in A. fumigatus compared to A. flavus. This may indicate that gpaB plays an essential role in the pathogenesis of A. fumigatus. G-proteins, as a main component of gpaB, have been shown to have a fundamental effect on transduction of the signals into the fungal cells.27,28 In fact, the activation of G-protein subunits in the cell wall of A. fumigatus increases cyclic adenosine monophosphate (cAMP) signaling, mitogen-activated protein kinase (MAPK) and protein kinase A (PKA) pathways in the fungi, and deletion of A. fumigatus GprC and GprD genes lead to growth destruction in all conditions, and an analysis of virulence revealed a significant decrease in virulence and delayed mortality in a murine model of aspergillosis.12 Down-regulation gpaB expression was detected mainly in COPD patients; the patients exhibited no expression of this gene. This finding is supported by the activation of other regulatory mechanisms, such as the MAPK signal pathway, ray proteins, histidine kinases, calcium signaling or signal transduction by other virulence subunits of G-proteins in the pathogenesis of IA.29
We observed the highest sidA gene expression in transplant recipients and low expression of this gene in cancer and COPD patients. It has been well-established that sidA plays a crucial role in virulence due to its high-affinity iron uptake systems in fungi, which triggers pathogenesis in the patients.30 Additionally, sidA genes excite Aspergillus growth in lavage fluid, also siderophore deficiency decreases tissue inflammation and invasion.31
Consistent with our finding, Gravelat et al. showed MedA accomplishes conidiation, because in ΔmedA strain of A. fumigatus, biofilm formation and adherence to pulmonary epithelial, endothelial cells and fibronectin have been impaired in vitro. The ΔmedA strain also had reduced the ability to damage pulmonary epithelial cells. Moreover, A. fumigatus ΔmedA strain displayed low virulence in both an invertebrate and a mammalian model of IA.32
The lack of a significant correlation between sidA expression and underlying diseases may be related to the activation of other systems for iron uptake by the Aspergillus species.33 The metaloreductases (MR) increased iron uptake upon the initiation of pulmonary IA infections in animal models.34,35
It would be beneficial for future studies to investigate sidA and gpaB gene expressions in other immunocompromised patients, such as HIV and hepatitis patients. In summary, the sidA and gpaB gene expression patterns in patients indicated that these genes play important roles in the pathogenicity of Aspergillus isolates; nevertheless, several regulatory genes and proteins appeared to be responsible for the unexpressed sidA and gpaB genes in the isolates, which explain the role of this main mechanism.
ConclusionTaken together, our findings indicated that the A. flavus was the major pathogen in cancer patients and transplant recipients. A high expression of gpaB in COPD samples and an increased expression of sidA in transplant recipient may be related to the critical role of predisposing factors in the expression of specific genes in our study population.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by a grant from Iran University of Medical Sciences, Tehran, Iran (Grant #25653).