The biodiversity and evolution of the microbial community in açai fruits (AF) between three geographical origins and two spontaneous decay conditions were examined by applying culture-independent methods. Culture-independent methods based on 16S rRNA from fifteen samples revealed that Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Acidobacteria were the most abundant phyla. At the genus level, Massilia (taxon with more than 50% of the sequences remaining constant during the 30h of decay), Pantoea, Naxibacter, Enterobacter, Raoultella and Klebsiella were identified, forming the carposphere bacterial microbiota of AF. AF is fibre-rich and Massilia bacteria could find a large quantity of substrate for its growth through cellulase production. Beta diversity showed that the quality parameters of AF (pH, soluble solids, titratable acidity and lipids) and elemental analysis (C, N, H and C/N ratio) were unable to drive microbial patterns in AF. This research offers new insight into the indigenous bacterial community composition on AF as a function of spontaneous postharvest decay.
Euterpe oleracea Martius is a palm tree that occurs all across the Amazon basin and is particularly abundant in the eastern Amazon. This species grows in floodplains, on land and in swampland soils. Fruiting occurs throughout the year, with a period of higher production from July to December. Açai fruits (AF) have a round shape with a diameter of 1–2cm and a weight of 0.8–2.3g. These fruits are composed of kernel (endosperm), which represents approximately 85–95% of the fruit volume. The mesocarp has a thickness of only 1–2mm, and the exocarp is a thin layer that is covered with a wax cuticle when ripe.1
Just prior to harvest, AF suffer a rupture at their apex, allowing access to microorganisms and oxygen. In addition, as AF do not have a thick exocarp for effective protection, they can be easily damaged during handling and transportation. Shipping occurs via small vessels across the Amazon estuary under optimal temperature (30°C), moisture (99%) and nutrient availability conditions for microbial growth. The transport time to the main trading centres in the region is between 8 and 30h, which is another important variable in this context.2 During transport, AF deterioration can occur in the holds of boats due to a lack of ventilation, causing nutritional and functional losses mainly through the activity of polyphenoloxidase, which can be easily followed by anthocyanin degradation.1
High levels of Total Mesophilic Bacteria (TMB) in the açai juice have been reported, reaching average values of 6logCFUg−1 dry matter (DM) and gaining a 1st and 2nd logarithmic order 11.3 and 29h after harvest.2 Initial values of TMB, acetic and lactic acid bacteria in AF of 6, 4, and 6.5logCFUg−1 fruit, respectively, were observed before suffering spontaneous decay.3 The isolation and identification of lactic acid bacteria in enrichment cultures from AF have been reported.4 However, the diversity and dynamics of microflora of AF were never investigated using high-throughput sequencing and quantification of bacterial diversity, to the best of the author's knowledge.
Recently, açai market began to be affected due to the occurrence of outbreaks of human infection by Trypanosoma cruzi, the protozoan responsible for Chagas disease. The native bacterial community present in AF is responsible for production of volatile organic compounds, detectable by triatomine (vectors) antennas and attracting them to the fruit.3,5
Metagenomic techniques provide insight for documenting the unexplored biodiversity and ecological characteristics of either whole communities or individual microbial taxa.6 Among AF, little is known about the bacterial communities that are involved in this habitat (e.g., high lipid and phenolic compound content), and in these conditions, the possibility of finding technologically promising species is high.
This study is the first to investigate the dynamics and diversity of the native bacterial community in AF during postharvest decay. The quantitative effects of quality parameters of juice on this microbial community were investigated using cultivation-independent approaches. Due to the microbiological contamination in the supply chain of AF, the scope will illustrate the potential function of these bacterial communities in the context of postharvest decay, field location and environmental conditions.
Materials and methodsStudy area and samplingAF were collected in three municipalities in the state of Pará (Brazil), i.e., Belém – Combu island (1°29′45.0″S 48°26′26.6″W), Abaetetuba – Campompema island (1°44′39.8″S 48°55′09.5″W) and Benevides – Benfica (1°17′45.7″S 48°17′41.0″W). The choice of these fruits was grounded in spatial variability and temporal heterogeneity; the AF from Benevides were of the ‘BRS-Pará’ variety developed by EMBRAPA (Belém, Brazil) and adapted for land areas.7 The fruits of other municipalities were native cultivars and were located in floodplain areas. AF from Abaetetuba were chosen because this municipality is part of the largest producer microregion in Brazil with 66,177tonnes (2014 data). Fruits from Belém were selected because of the proximity to the laboratory. AF were collected in October-November 2013. 50kg of fruits were collected from each location and transported under refrigeration (10±2°C) to the laboratory. The total time between harvest and the beginning of the experiment was 0.5h (Combu island), 2h (Benfica) and 5h (Campompema island). To assess the potential variation in the bacterial community in AF arising from environmental heterogeneity and to reduce bias for replication, the total mass of AF was obtained from 11 to 23 bunches (two bunches per tree with height of 15±5m) with 2–4kg of fruits per bunch. The maturity stages of the fruits ranged between 9 and 11 according to Rogez et al.8 classification.
Decay conditions and sample preparationThe two most common conditions of AF transport on boats were simulated as described by Aguiar et al.3 Briefly, 2kg of AF from each sample collection was taken from 50kg of fruits by quartering and added to four closed polystyrene boxes (anaerobic condition, such as in boat cargo holds) and to four open baskets (aerobic condition, such as on the open decks of boats). Under both aerobic and anaerobic conditions, AF from each municipality were collected at times 0 (starting point in the laboratory, generating three samples), 10.0h (a total of six samples) and 30.0h (a total of six samples). The parameters of temperature and relative humidity during the postharvest period of AF from Combu island, Campompema island and Benfica in anaerobic conditions were 27.8±0.9°C and 45±7.5%, 28±0.9°C and 38±4.9%, 28.1±0.6°C and 66±3.0%, respectively. The same parameters, for the aerobic condition were 28.1±0.8°C and 48±5.1%, 26.9±1.5°C and 55±4.1%, 28.9±1.8°C and 68±5.9%, respectively.
The pulping was performed according to the traditional juice preparation method.2 In order to characterize the native microbiota of AF, no thermal treatment (blanching and/or pasteurization) was performed. To date, Brazilian law does not oblige small and large industries to perform thermal treatments in the preparation of juice or pulp. Therefore, the pulping employed in this study is the same from that carried out industrially.
Weight loss and fruit temperatureThis parameter was monitored by the gravimetric method. Under both aerobic and anaerobic conditions, AF were weighed in synthetic nets in 2.0kg portions at each sampling time. The temperature of the AF was monitored using thermometers (±0.2°C) located at the centre of the stack of fruits.
Respiration ratesThe respiration rate was measured in AF according to Aguiar et al.3 The percentages of O2 and CO2 in the pots were obtained using a gas analyser (PBI Dansensor; Checkpoint II Portable Gas Analyser, Ringsted, Denmark). The respiration rate of O2 and CO2 (mLkg−1h−1) was converted to mmolkg−1h−1 using the ideal gas law.
Quality parametersAn analysis of quality parameters, such as soluble solids (°Brix), total solids (TS), pH, titratable acidity (TTA) and lipids, was performed after processing. All of the analyses were based on the AOAC method and carried out in triplicate.9
Elemental analysisAn elemental analysis of carbon (C), hydrogen (H) and nitrogen (N) was performed using the instrument LECO® TruSpec CHN (Leco Corp., St. Joseph, MI). Approximately 0.10g of dry açai juice sample was weighed in tin foil (Leco®) and fired at 950°C. The test was performed in triplicate and expressed as a percentage of C, H and N on a dry basis (w/w).
DNA extraction and PCRThe total DNA was extracted from 1.5mL of açai juice that was fixed (1:1 v/v) in RNA later (Ambion, Life Technologies, Carlsbad, CA) using PowerSoil® DNA Isolation Kits acoording to manufacture instructions (MO BIO Laboratories, Carlsbad, CA). The total DNA served as the template for the amplification of the V4 region of the bacterial 16S rRNA gene (average sequence length ∼450bp) in the polymerase chain reaction using primers Bakt_341F/Bakt_805R (5′-CCTAC GGGNGGCWGCAG-3′,5′-GACTACHVGGGTATCTAATCC-3′) and barcodes that were attached to adapters according to the manufacturer's protocol (Life Technologies). The PCR was carried out in a final volume of 50μL containing Buffer 1×, dNTP 0.2mmolL−1, MgCl2 4.0mmolL−1, 0.5μmolL−1 of each primer, 0.7U of Taq DNA polymerase and 0.3mgmL−1 of Bovine Serum Albumin. The cycling conditions consisted of an initial denaturation step at 95°C for 5min, and two cycles composed of 95°C for 60s, 48°C for 60s and 72°C for 60s, two cycles composed of 95°C for 60s, 50°C for 60s and 72°C for 60s, two cycles composed of 95°C for 60s, 52°C for 60s and 72°C for 60s, 22 cycles composed of 95°C for 60s, 54°C for 60s and 72°C for 60s.
Sequencing, diversity and statistical analysisEquivalent amounts of each sample were added to the reaction mixture for sequencing and there was no replicate analysis for any sample. The libraries were sequenced in Ion PGMTM using an Ion PGMTM Sequencing 400 Kit and finally deposited into two Ion 318TM chip Kits v2 according to the manufacturer's protocol (all kits and systems were provided by Life Technologies).
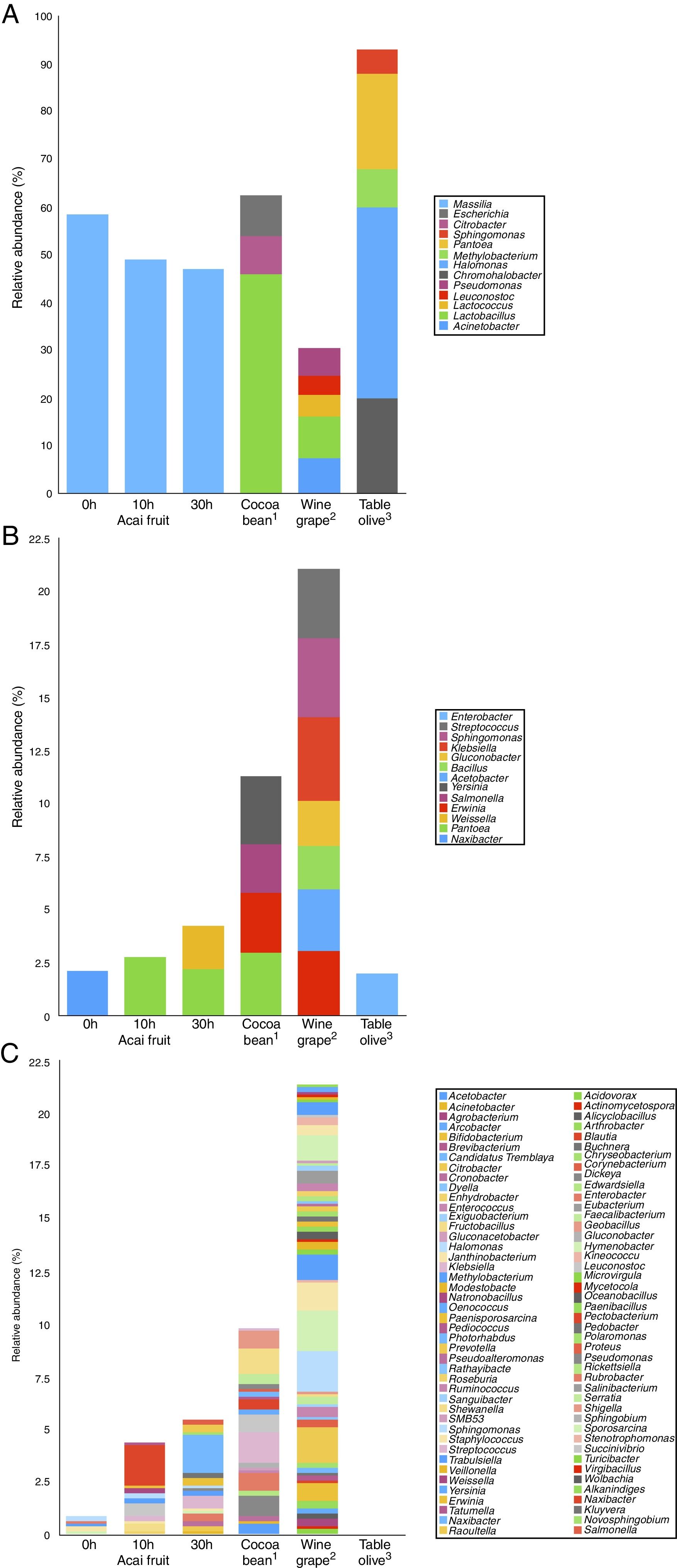
The data analyses were performed using QIIME10 and USEARCH.11 First, raw data (reads) with low quality (maximum error probability of 0.5), smaller than 200bp and considered chimaeras were eliminated. Then, samples were demultiplexed according to barcode sequence for OTU clustering, using QIIME with UCLUST method.11 The alpha and beta diversity (rarefied samples) analyses were calculated with QIIME and statistical analyses, e.g., redundancy analysis (RDA) and Principal coordinates analysis (PCoA), were done with R-project (http://www.R-project.org/), using BiodiversityR12 and Vegan13 libraries. A graphical comparison of relative abundance (percentage of bacterial community at the classification level of genus) was made with three usual foods well known for their spontaneous fermentation process or decay (wine grape14; table olives15 and cocoa bean16). These comparisons were graphically achieved without any bioinformatics treatment on the data.
Overall schemeIn this investigation, we performed comprehensive study of the biodiversity and evolution of the microbial community in AF between three geographical origins during the spontaneous decay conditions (aerobic and anaerobic). A general scheme presenting the entire experimental design with information about the study area, conditions of collection, preparation of the samples and developed analyses is presented in Fig. S1.
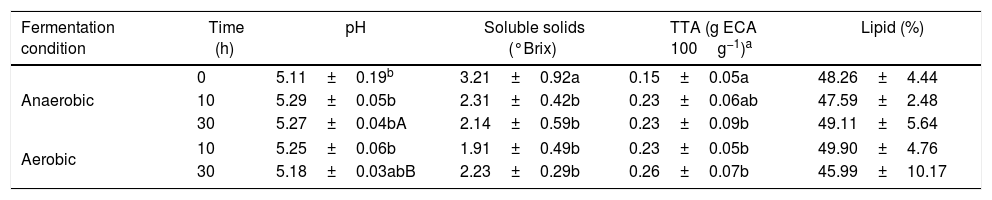
ResultsQuality parameters during the spontaneous decay of AFThe quality parameters from the açai juice that was produced by AF throughout the postharvest time under anaerobic and aerobic conditions are presented in Table 1. For each condition of decay, all of the properties showed no significant variation during the postharvest time, with the exception of pH value after 30h. Analysing the separate decay conditions for AF from anaerobic and aerobic conditions during postharvest decay, both systems presented significant variation (p<0.05) for pH and TTA, increasing with time and decreasing soluble solids. For all of the AF samples, the average TS after pulping was 9.05±1.81%.
Quality parameters of E. oleracea fruits during postharvest fermentation.
| Fermentation condition | Time (h) | pH | Soluble solids (°Brix) | TTA (g ECA 100g−1)a | Lipid (%) |
|---|---|---|---|---|---|
| Anaerobic | 0 | 5.11±0.19b | 3.21±0.92a | 0.15±0.05a | 48.26±4.44 |
| 10 | 5.29±0.05b | 2.31±0.42b | 0.23±0.06ab | 47.59±2.48 | |
| 30 | 5.27±0.04bA | 2.14±0.59b | 0.23±0.09b | 49.11±5.64 | |
| Aerobic | 10 | 5.25±0.06b | 1.91±0.49b | 0.23±0.05b | 49.90±4.76 |
| 30 | 5.18±0.03abB | 2.23±0.29b | 0.26±0.07b | 45.99±10.17 | |
Under anaerobic conditions, there was no variation in weight loss during experimentation. Under aerobic conditions, the mean weight loss was 0.94±0.53%, 0.66±0.48% and 0.70±0.58% for AF from Combu island, Campompema island and Benfica, respectively. Under both conditions, the minimum temperature was 25.0°C, and the maximum temperatures were 32.2°C and 32.6°C in the aerobic and anaerobic conditions, respectively, during 30h of experimentation (Fig. S2).
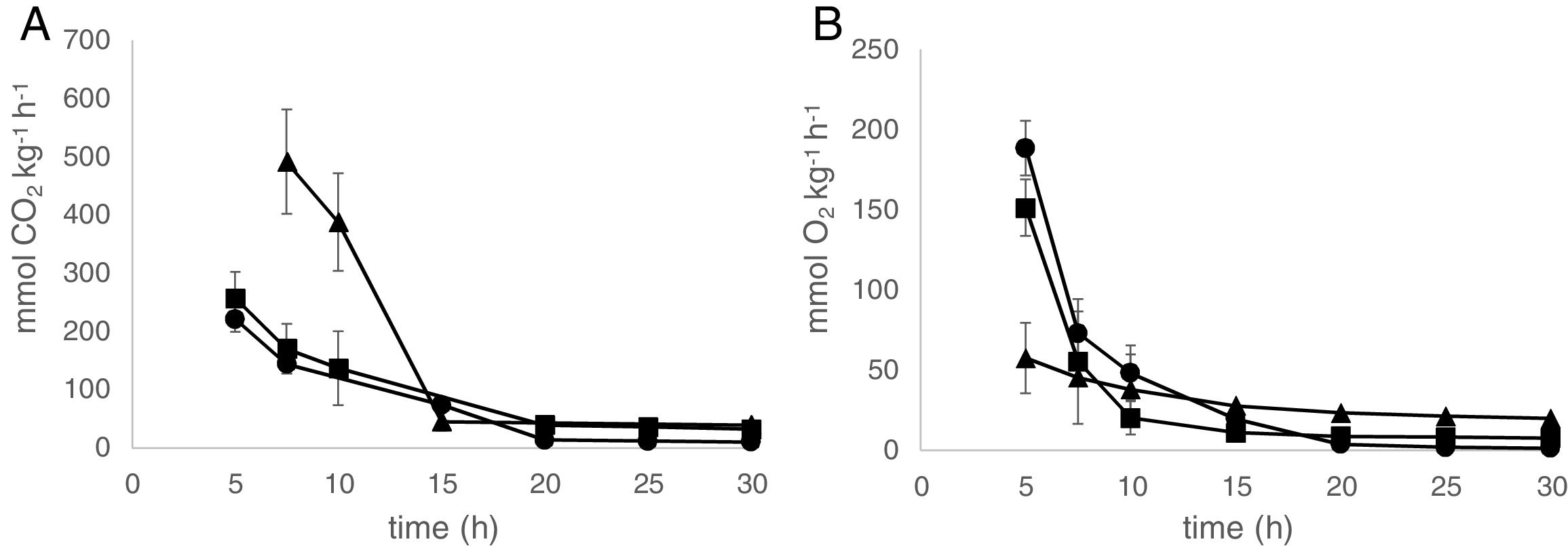
Respiration ratesFig. 1 represents the respiration rates of AF from three municipalities under anaerobic conditions of decay. AF from Combu and Campompema islands showed similar values during postharvest decay. The Benfica locality showed higher values during the first 15h compared with Combu and Campompema islands. The initial respiration rates under anaerobic conditions for CO2 production were 220.97±22.41mmolkg−1h−1, 257.15±45.04mmolkg−1h−1 and 491.45±89.94mmolkg−1h−1 for Combu island, Campompema island and Benfica, respectively. For all municipalities, the CO2 production rate decreased to less than 100mmolkg−1h−1 (39.1±10.15%) after 15h until the end decay (Fig. 1A).
The respiration rates for O2 absorption presented the same behaviour. The initial values from Combu island, Campompema island and Benfica were 188.57±16.93mmolkg−1h−1, 151.27±17.60mmolkg−1h−1 and 57.62±22.00mmolkg−1h−1, respectively. A strong consumption of O2 could be observed during the first 10h for AF from Combu island (48.36±16.93mmolkg−1h−1) and Campompema island (20.24±10.31mmolkg−1h−1), reaching a percentage of 2.5±0.8% (Fig. 1B).
Elemental analysisThe mass fractions of C, H, N and C/N ratio parameters from açai juice that was produced by AF during the postharvest time are presented (Table S1). The data showed no significant difference from the gross composition in the storage conditions and postharvest time of AF.
Bacterial diversity in the postharvest decay of AFWe analyzed a total of 10,507,497 reads (Table S2). After filtering the reads, 770,247 bacterial 16S rRNA sequences were obtained, ranging from 13,285 to 96,870 in all 15 samples of AF. In total, 4092 OTUs were assigned based on a 3% dissimilarity threshold, with 1670 unique OTUs. Good's coverage estimate was calculated to assess the percentage of captured diversity by the devoted sequencing effort, and our results demonstrated that the sequencing efforts were satisfactory, with 0.87 on average and varying from 0.74 to 0.92. The microbial core among postharvest decay per sample, Combu island (Table S3), Campompema island (Table S4) and Benfica (Table S5) and the microbial core across 100% of samples (Table S6) were presented. The sequencing data of the 16S rRNA genes are publicly available in the NCBI Short Read Archive (SRA) under accession no. PRJNA284888.
Based on the USEARCH and QIIME analysis, the majority of these sequences for all AF could be affiliated with five phyla: Proteobacteria (>90%), Firmicutes (2%), Actinobacteria (0.2%), Bacteroidetes (0.1%) and Acidobacteria (0.05%). With approximately 200 bacterial genera identified, the six most abundantly represented were Massilia (>50%), Pantoea (3%), Naxibacter (2%) and Enterobacter, Raoultella and Klebsiella (<1% each). The bacteria that increased during AF postharvest decay were members of the family Enterobacteriaceae (mostly under anaerobic conditions), highlighting the following genera: Pantoea (3%), Klebsiella (1%), Enterobacter, Erwinia, Buttiauxella, Cedecea, Citrobacter, Cronobacter, Kluyvera, Salmonella, Serratia, Tatumella, Raoultella, Gibbsiella, Leclercia and Mangrovibacter (< 1% each). Likewise, the families Leuconostocaceae (Fructobacillus, Leuconostoc and Weissella), Streptococcaceae (Lactococcus) and Microbacteriaceae (Curtobacterium) showed the same increase, mostly after 10h. Thus, we concluded that the main bacteria from AF were gram-negative organisms.
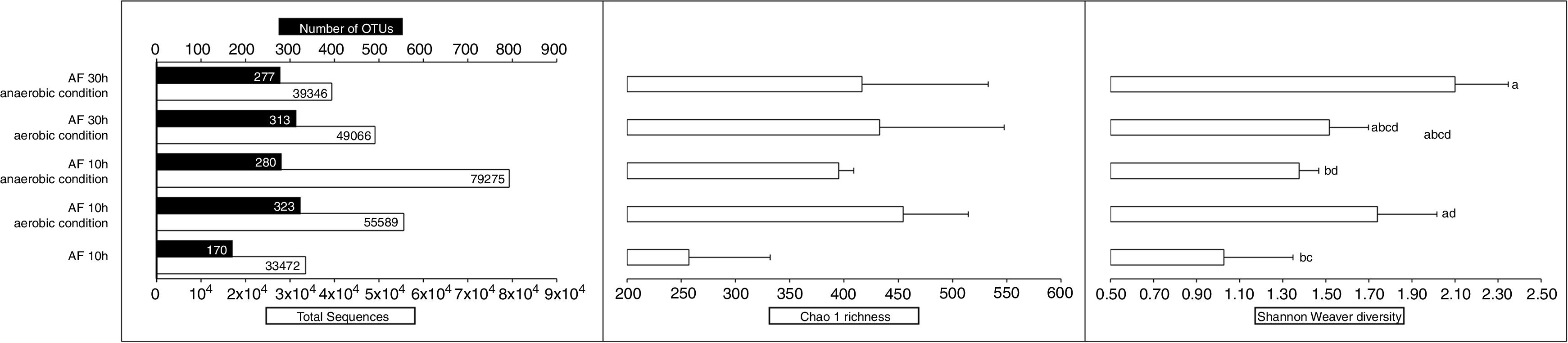
The richness, diversity estimation and bacterial community composition of AF were compared by rarefaction analysis, Chao1 and Shannon-Weaver (Fig. 2) and weighted UniFrac algorithm, respectively. Rarefaction curves indicated that the entire richness of the bacterial communities were not captured, as the curves did not reach a plateau with increasing sample size, and more sequencing efforts were needed. The rarefaction curves, that were obtained with 11,739 to 56,956 sequences, revealed no significant differences between the AF samples for the lowest number of sequences. However, for samples with greater coverage (AF at 10h with spontaneous decay under both aerobic and anaerobic conditions), there was a significant difference (p<0.05) between these samples. These results suggest a higher complexity of the bacterial community in AF during postharvest decay.
Overview of the total sequences, number of OTUs and microbial diversity of E. oleracea fruits during postharvest fermentation under both aerobic and anaerobic conditions. Ecological measures were indicated by Chao1 richness estimator and Shannon-Weaver diversity index. The calculation of the richness estimator and diversity index was based on OTU tables rarefied. The total sequences refer to the total number of taxonomically assigned sequences, and OTUs were defined as <3% nucleotide sequence difference (n=3). Values with different letters are significantly different (p<0.05), as deduced from the Tukey test. AF=Açai fruit.
Based on the Chao1 richness estimation there was no significant variation between AF samples. The coverage of our experiments was predicted, with a total richness ranging from 49.4% (AF from Campompema island) to 76.36% (AF from Combu island). This coverage indicates a good sampling (spatial variability and temporal heterogeneity) of AF and a high-throughput sequencing method. The library construction could represent most of the species in the AF carposphere.
The Shannon-Weaver diversity index indicates a significant difference (p<0.05) in the diversity between AF during postharvest decay. The highest value was observed in AF after 30h under anaerobic condition.
The analysis of phylogenetic distance on bacterial communities using hierarchical clustering demonstrated minimal differences in the community structure in AF when comparing all fifteen samples. Our results revealed little distinct bacterial community variation in bacterial richness across the spontaneous decay of AF at 30h. The weighted UniFrac distance reinforced this suggestion because, considering both the similarity between sequences and the sequences abundances, our data demonstrated minimal differences in community structure in AF when comparing all fifteen samples.
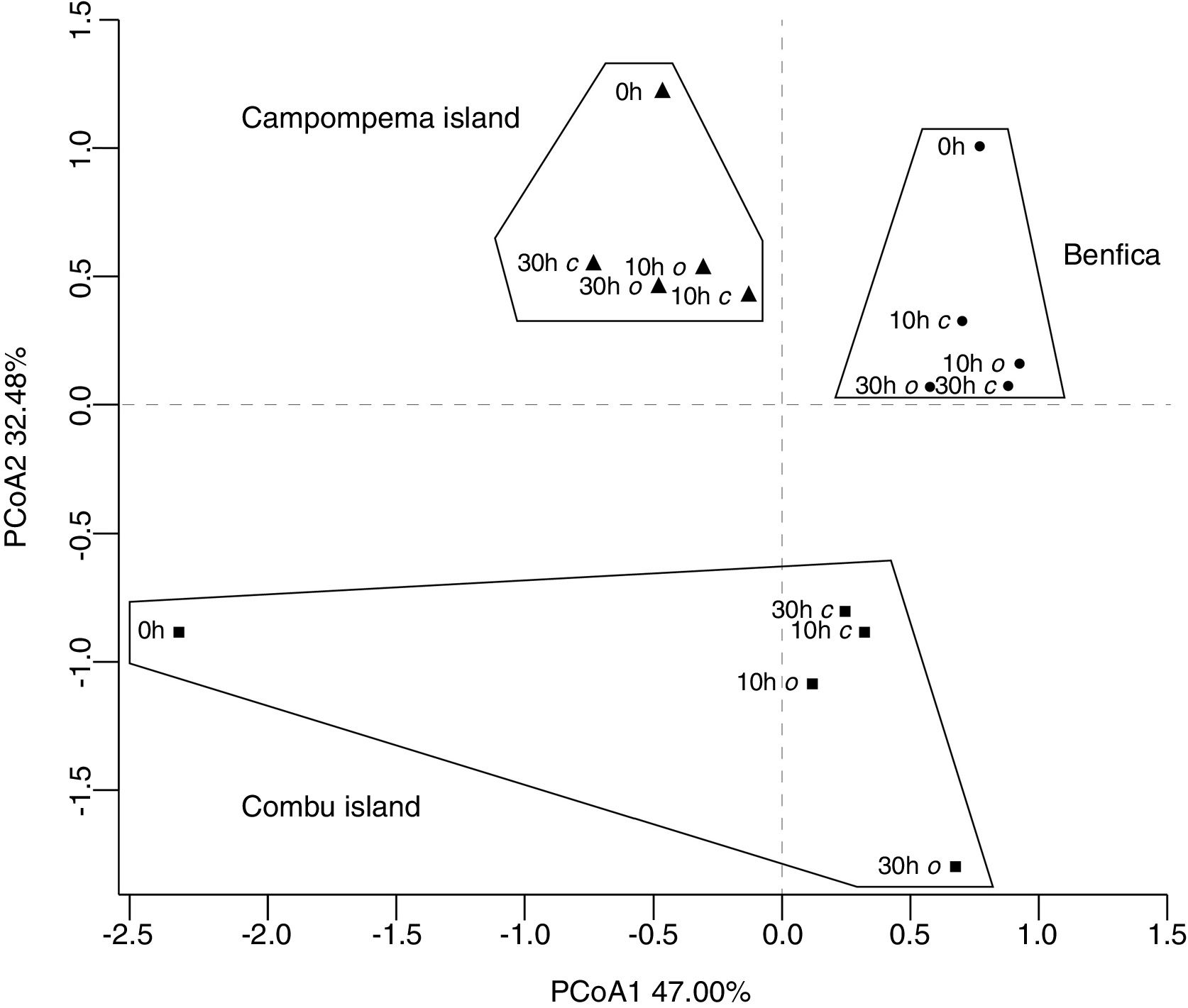
The relationship between bacterial communities linkage to the environmental context dataPCoA based on OTU composition was used for the overall comparison of significant differences among the bacterial communities by finding clusters of samples that will reflect the similarity of the biological communities. The first two principal components could explain 79.48% of the variation of the total bacterial community in the AF samples (Fig. 3). The PCoA analysis could not explain the type of decay conditions that were applied to AF; however, there was a clustering regarding the sampling locations. In addition, the samples in the beginning of the experiment were not grouped with samples at 10h and 30h for all locations. The bacterial community of the initial AF from Combu island was well separated from the all of the other samples along the first component (PCoA1), and the set of all samples from Benfica and Campompema island was separated from the samples from Combu island along the second component (PCoA2).
Principal coordinates analysis plot illustrating the weighted UniFrac distances between bacterial communities in samples of açai fruit under two postharvest conditions (anaerobic – c and aerobic – o). Açai fruits were sampled from three localities of state of Pará. Stars: Combu island, squares: Campompema island, and circles: Benfica.
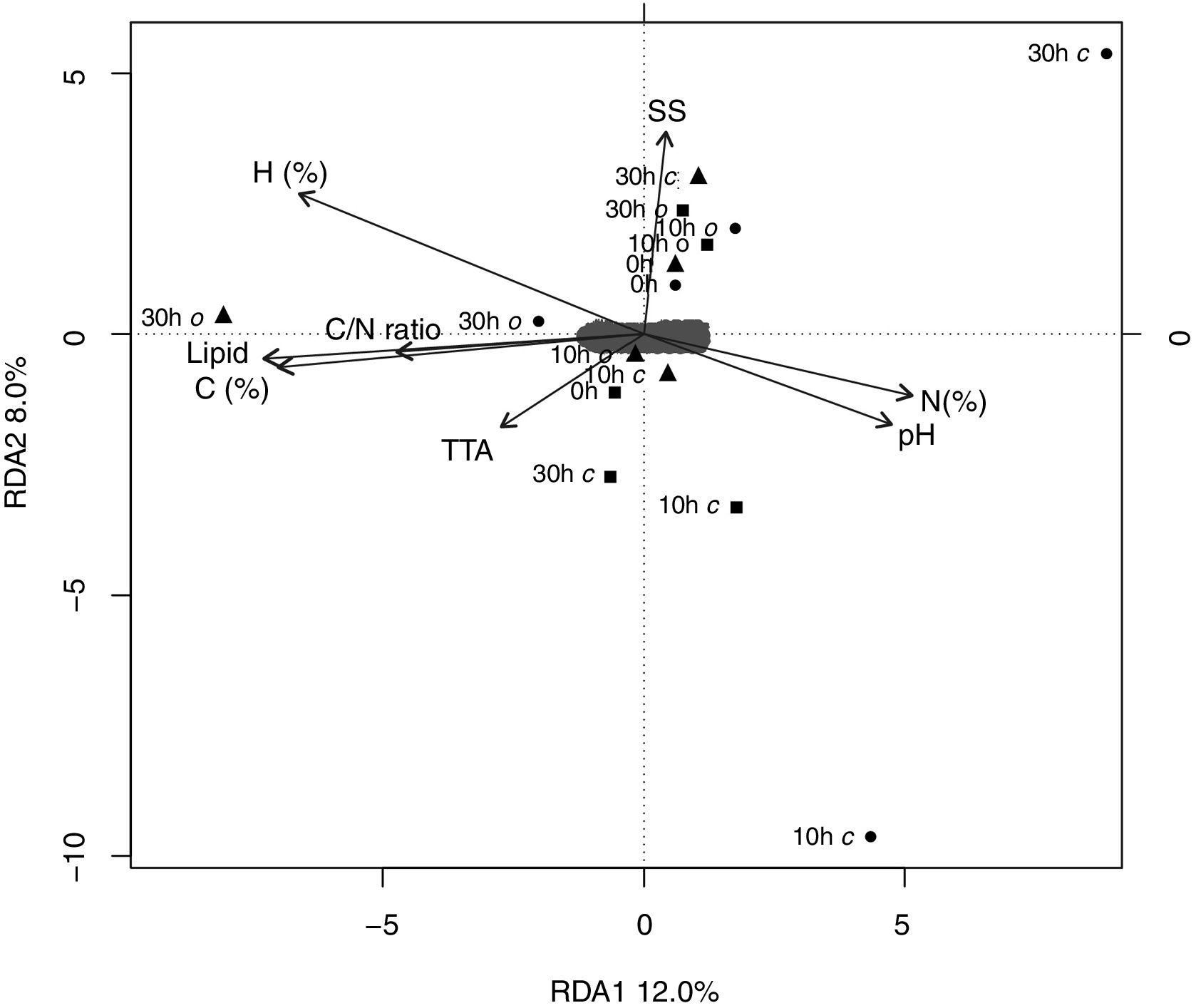
The RDA was performed on the OTU composition data and the quality parameters of AF showed that the first two RDA components could explain only 20.0% of the total variation (Fig. 4). The overall comparison would obscure the impact of some quality parameters of AF with the bacterial community.
Redundancy analysis (RDA) of the abundant taxon and açai fruit quality parameters of fifteen samples of açai fruits under two postharvest decay conditions (anaerobic – c and aerobic – o). Açai fruits were sampled from three localities of state of Pará. Squares: Combu island, triangles: Campompema island and circles: Benfica. Correlations are represented by the length and angle of arrows (environmental factor vectors).
The quality parameters and the elemental analysis from AF juice did not show significant difference (p<0.05) in function of decay condition for most studied parameters. Low acidic pH in AF encouraged bacterial fermentation to convert the major carbohydrates of the fruit to organic acids. The carbohydrate consumption during postharvest AF decay, and after 27h, the consumption of d-glucose and d-fructose by microorganisms was 73 and 95%, respectively.3
The carbohydrate consumption and the increase in TTA occurred for all samples. Lactic and acetic acid in AF following a first-order model, achieving values of 0.97 and 1.50mmolkg−1, respectively.3 Spontaneous decay typically results from the competitive activities of a variety of autochthonous and contaminating microorganisms. Those organisms that are best adapted to the conditions during the decay process will eventually dominate.17
The most representative phyla in AF were Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Acidobacteria. Other studies have used culture-independent and culture-dependent techniques to describe taxon abundances established in grapes (Firmicutes, Actinobacteria and Proteobacteria)14,18–20 and table olives (Proteobacteria and Firmicutes).21
We observed little distinct bacterial community variation between aerobic and anaerobic conditions of the spontaneous decay of AF. The increased families Enterobacteriaceae (Pantoea, Klebsiella, Enterobacter, Erwinia, Buttiauxella, Cedecea, Citrobacter, Cronobacter, Kluyvera, Salmonella, Serratia, Tatumella, Raoultella, Gibbsiella, Leclercia and Mangrovibacter), Leuconostocaceae (Fructobacillus, Leuconostoc and Weissella), Streptococcaceae (Lactococcus) and Microbacteriaceae (Curtobacterium) showed the same behaviour as in wine fermentation.18
The six most abundant bacterial genera that were identified in AF were Massilia (taxon remaining constant during the 30h of spontaneous decay), Pantoea (taxon with the highest increase during decay, especially after the first 10h), Naxibacter, Enterobacter, Raoultella and Klebsiella. The bacterial community structure in botrytized wine exhibited little change during fermentation, except for a gradual reduction of Proteobacteria and an increase of Firmicutes over time.18 Lucena-Padrós et al.21 studying the microbiota associated with Spanish-style green olives during fermentation, observed that only two species (P. agglomerans and P. ethanolidurans) could be detected during the final stage of fermentation (69–72d).
Massilia was isolated by culture-independent and culture-dependent techniques in many environmental samples, e.g., agricultural aerosols,22 apple phyllosphere,23 cucumber root and seed,24 leaf surface of lettuce22 and grape berry.20Massilia exhibited in vitro attributes related to plant growth promotion, including indole acetic acid and siderophore production. Curiously, another study related the production of extracellular enzymes by Massilia, such as chitinases and cellulases.25 Bichara and Rogez1 reported that AF is fibre-rich (15.85g 100g−1 DM), presenting a soluble/insoluble dietary fibre ratio of 1:3. Massilia bacteria could find a large quantity of substrate for its growth through cellulase production. Their high cellulosic, phenolic and lipid contents make this habitat (carposphere) helpful for the maintenance of the genus Massilia during 30h of spontaneous decay.
Pantoea has been associated with leaf spot necrosis in beach peas, onions and okra.26 A high relative abundance of Pantoea on surfaces of fresh lettuce (8.9%), spinach (32.4%), sprouts (bean) (57.5%), pepper (11.1%) and strawberries (10.4%) were exhibited.19 However, the genera Pantoea, Enterobacter and Klebsiella have been reported as nitrogen-fixing bacteria, PHB producers and phosphate solubilizers.27,28 A peculiarity of the AF is their high protein content (6.7–10.5g 100g−1 DM),1 which classifies this fruit as a rich tropical fruit.
Analyses of beta-diversity indicated the fifteen samples of AF that were harvested from areas with farming practices grown in dry land and others from native culture grown in low floodplain and under different conditions of spontaneous decay during the postharvest have very similar microbiomes. In addition, the differences in respiration rates and mass loss of AF did not significantly influence the bacterial diversity. Other environmental factors that could suggest significant relationships to drive microbial patterns in AF were analyzed by PCoA and RDA. PCoA showed that a cluster can be formed from AF location; however, all environmental context data that were analyzed in this study were not sufficient to establish clustering. In addition, RDA analysis confirmed that none of the environmental factors that were proposed in this study were capable of modulating the bacterial community in AF during postharvest time.
Due to the high content of phenolic compounds and lipids in AF, we compared the relative abundance of bacterial communities in food samples that are rich in these constituents (e.g., table olive, cocoa bean and wine grape) and also undergo a fermentation process. The genus-level of taxonomic resolution was achieved by next-generation sequencing (NGS) approaches and is presented in Fig. 5. The characterization of the bacterial diversity in AF showed similarities with phenolic- and/or lipid-rich fruits of the genera Pantoea, Enterobacter, Klebsiella, Citrobacter, Cronobacter, Salmonella, Erwinia, Acinetobacter, Enhydrobacter, Sphingomonas, Hymenobacter, Methylobacterium, Pseudomonas, Janthinobacterium, Leuconostoc, Weissella and Fructobacillus. Overall, there were high relative abundances of Enterobacteriaceae to colonize certain fruits and vegetables, but it is difficult to determine the specific factors that are responsible for driving the divergence between the bacterial diversity of different food samples.19 Likewise, the highest specific microbial taxa on AF (Massilia) cocoa bean (Lactobacillus), wine grape (Lactococcus) and table olive (Halomonas) are not the same. Despite the high biodiversity that was observed in wine grape, AF and cocoa bean compared with that in table olive, there is still a large gap in the knowledge of the functional diversity and significance of microbial interaction on these food samples. We addressed these knowledge gaps with metatranscriptomic studies, which not only have different abundances of bacterial taxa but also have different transcription levels of every gene across samples.
Relative abundance (%) of bacterial community on the genus rank of (a) major (4–100%), (b) intermediate (2–3.99%) and (c) lower (0.01–1.99%) as obtained by Next-Generation Sequencing from Açai fruit, cocoa bean, wine grape and table olive. 1Based on experimental data from Illeghems et al.16; 2Based on experimental data from Bokulich et al.14; 3Based on experimental data from Cocolin et al.15
This study served as a pilot study using NGS to profile the bacterial community structure in AF. NGS methods have fundamentally enhanced our understanding of microbially dominated ecosystems and revealed a number of new microbial types that have no cultured relatives and have potential technological applications. The AF can be used as a source for isolating polyhydroxyalkanoates-producing bacteria (e.g., Massilia sp.), which are attractive candidates for use in the production of biopolymers from renewable carbon sources.
Conflicts of interestThe authors declare no conflict of interest.
The authors are grateful to Fundação de Amparo à Pesquisa do Estado do Pará (FAPESPA), Pró-Reitoria de Pesquisa e Pós-Graduação (PROPESP/UFPA), Fundação de Amparo e Desenvolvimento da Pesquisa (FADESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), all from Brazil, for their financial support.
The following are the supplementary data to this article: