Repeated application of pesticides disturbs microbial communities and cause dysfunctions on soil biological processes. Granstar® 75 DF is one of the most used sulfonylurea herbicides on cereal crops; it contains 75% of tribenuron-methyl. Assessing the changes on soil microbiota, particularly on the most abundant bacterial groups, will be a useful approach to determine the impact of Granstar® herbicide. For this purpose, we analyzed Actinobacteria, which are known for their diversity, abundance, and aptitude to resist to xenobiotic substances. Using a selective medium for Actinobacteria, 42 strains were isolated from both untreated and Granstar® treated soils. The number of isolates recovered from the treated agricultural soil was fewer than that isolated from the corresponding untreated soil, suggesting a negative effect of Granstar® herbicide on Actinobacteria community. Even so, the number of strains isolated from untreated and treated forest soil was quite similar. Among the isolates, resistant strains, tolerating high doses of Granstar® ranging from 0.3 to 0.6% (v/v), were obtained. The two most resistant strains (SRK12 and SRK17) were isolated from treated soils showing the importance of prior exposure to herbicides for bacterial adaptation. SRK12 and SRK17 strains showed different morphological features. The phylogenetic analysis, based on 16S rRNA gene sequencing, clustered the SRK12 strain with four Streptomyces type strains (S. vinaceusdrappus, S. mutabilis, S. ghanaensis and S. enissocaesilis), while SRK17 strain was closely related to Streptomyces africanus. Both strains were unable to grow on tribenuron methyl as unique source of carbon, despite its advanced dissipation. On the other hand, when glucose was added to tribenuron methyl, the bacterial development was evident with even an improvement of the tribenuron methyl degradation. In all cases, as tribenuron methyl disappeared, two compounds were detected with increased concentrations. These by-products appeared to be persistent and were not degraded either chemically or by the studied strains. Based on these observations, we suggested that bacterial activity on carbon substrates could be directly involved in the partial breakdown of tribenuron methyl, by generating the required acidity for the first step of the hydrolysis. Such a process would be interesting to consider in bioremediation of neutral and alkaline tribenuron methyl-polluted soils.
The soil ecosystem is the theater of complex linked biological processes, regulated by its microbiota. Changes, such as introducing xenobiotic substances, cause fluctuation on microbial quality and quantity, affecting then the soil balance. Therefore, the microbial population reflects environmental changes and may be taken as an efficient indicator to evaluate the impact of exogenous molecules.1
Sulfonylurea herbicides are widely used around the world to protect cereal crops. They could constitute a long-term environmental hazard,2 with even a contamination risk of groundwater and surface waters, due to leaching processes.3,4 Furthermore, they are considered as disturbers for the soil microbiota.1,5 Granstar® 75 DF (DuPont de Nemours) is one of the most used herbicides in Algeria.6 It is used under different commercial denominations, in North Africa, Europe, North and South America and Asia.7–10 This herbicide contains 75% of tribenuron-methyl (TBM), a sulfonylurea molecule active against a large number of annual dicotyledons11 and it is used in low but effective concentrations (12g/300L/ha). TBM acts by stopping cellular division of meristematic tissues of plants via inhibition of acetolactate synthetase; an enzyme present in higher plants, bacteria and fungi, but absent in humans and animals.7,12,13
Several studies reported effects of sulfonylurea herbicides on global microbial community and on specific groups like fungi.1,5,13 However, the impact on soil Actinobacteria is still little explored, despite their abundance and importance in soil.14 Actually, these Gram-positive bacteria have considerable potential for biotransformation and biodegradation of pesticides,15,16 due to their ability to produce variable extracellular enzymes, degrading complex and recalcitrant pollutants17 in various environments.18
The main objective of this study was to examine the effect of Granstar® herbicide on soil Actinobacteria. For that purpose, Actinobacteria strains were isolated from both untreated and Granstar® treated soils. The resistance of isolated strains to different concentrations of the herbicide was evaluated. The two most resistant strains were further characterized and the impact of TBM on their growth determined.
Materials and methodsSoil treatment and Actinobacteria isolationHerbicide Granstar® 75 DF was obtained as a commercial powder from Du Pont de Nemours (Algeria). Three soils were selected for our study. Two agricultural soils, from two different eastern regions of Algeria: Ain Karma, Constantine (soil 1) and Ain Babouche, Oum El Bouaghi (soil 2). The third soil was from a forest, taken at Chaabet Ersas, Constantine (soil 3). Soils 2 and 3 were free of pesticides, while soil 1 was regularly treated with various herbicides, including Granstar®. Soil samples were collected at 10cm depth. The soils 2 and 3 were incubated at laboratory with the herbicide as follows: Granstar® was added to 150g of soil at 40mgL−1, which is the recommended field application dose. The incubation was performed in covered aerated two liters beakers at room temperature for three weeks. Hereafter, these treated soils are named treated soil 2 and treated soil 3.
For isolation of Actinobacteria strains, 10g of each soil sample (soil 1, soil 2, soil 3, treated soil 2, treated soil 3) were suspended in 90mL of physiological water (NaCl 9‰). For platting isolation, serial dilutions (10−2, 10−3 and 10−4) were prepared using physiological water. Isolation was carried out on Bennett agar, containing 10g glucose, 2g casamino acid, 1g meat extract, 1g yeast extract, 15g agar per liter of distilled water, pH 7.3,19 supplemented with 40mgL−1 of Granstar®. This medium allows Actinobacteria growth20 and is considered as a production medium for enzymes and bioactive substances.21 After incubation at 30°C for a week, Actinobacteria colonies were selected by a direct light microscopic observation (×10) based on their morphological characteristics (presence of short branching filaments). The isolated strains were preserved on 50% glycerol solution (v/v) at −80°C and on starch-casein agar slant, after sporulation, at 4°C.
Herbicide-resistance screeningResistance of the isolated strains to various doses of Granstar® was verified by using Bennett agar medium supplemented with herbicide at different concentrations: 0.004, 0.04, 0.05, 0.075, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5, 0.55, 0.6 and 0.65% (v/v). The plates were incubated at 30°C for 7 days.
Morphological and cultural characterization of strainsFour ISP (International Streptomyces Project) media; ISP2 (yeast extract 4g, malt extract 10g, glucose 4g, agar 20g, distilled water 1L, pH 7.3), ISP4 (soluble starch 10g, NaCl 1g, CaCO3 2g, K2HPO4 1g, MgSO4·7H2O 1g, (NH4)2 SO4 1g, trace salts solution [FeSO4·7H2O 1g, ZnSO4·7H2O 1g, MnCl2·4H2O 1g, distilled water 1L, pH 7.0] 1mL, agar 20g, distilled water 1L, pH 7.0–7.4), ISP6 (peptone 15g, proteose peptone 5g, ferric ammonium citrate 0.5g, sodium thiosulphate 0.08g, yeast extract 1g, K2HPO4 1g, agar 20g, distilled water 1L, pH 7.0–7.2), ISP7 (glycerol 15g, l-asparagine 1g, K2HPO4 0.5g, NaCl 0.5g, FeSO4·7H2O 0.01g, agar 20g, distilled water 1L, pH 7.2–7.4), as well as starch-casein agar (soluble starch 10g, casein 1g, K2HPO4 0,5g, agar 20g, distilled water 1L, pH 7.0–7.5) were inoculated to record morphological and cultural features of the selected resistant strains.22 Sporulation and fragmentation of the substrate mycelium, as well as the characteristics of produced spores on aerial mycelium were determined by the lamella technique,22 which consists in inserting a sterile lamella on the surface of ISP2 agar, at an angle of 45°, the inoculum was then deposed on the lamella, in contact with the surface of the medium. After 14 days of incubation, the lamella was gently removed, deposed on a glass slide and observed using a light microscopy oil immersion objective. Details about aerial mycelium color and production of diffusible pigments and melanin were obtained by inoculating ISP4, starch-casein, ISP6 and ISP7 agars, respectively.22,23
Phylogenetic analysesThe DNA extraction of the isolates was performed from a 48h liquid culture in Luria-Bertani (LB) broth by using the Ultraclean Microbial DNA Isolation kit (Mo Bio). For the amplification of the 16S rRNA genes, the bacterial primer sets 8F24 and 1387R25 were used with a program of an initial denaturation step at 95°C for 10min, followed by 35 cycles of 45s at 95°C, 45s at 56°C and 1min at 72°C, and a final extension at 72°C for 10min, in an AB Applied Biosystems Veriti 96 well thermal cycler as previously described.26 The purification of the amplified DNA was performed with the GFX PCR DNA and the Gel Band Purification kit (GE Healthcare) as described.27 DNA sequences were obtained by GATC Biotech AG (Germany) using the Sanger method.28 The sequences were corrected using the Sequencher v. 4.1.4 software (Gene Codes) and subsequently deposited with Genbank under accession numbers KR871404 and KR871405 for SRK12 and SRK17 sequences, respectively.
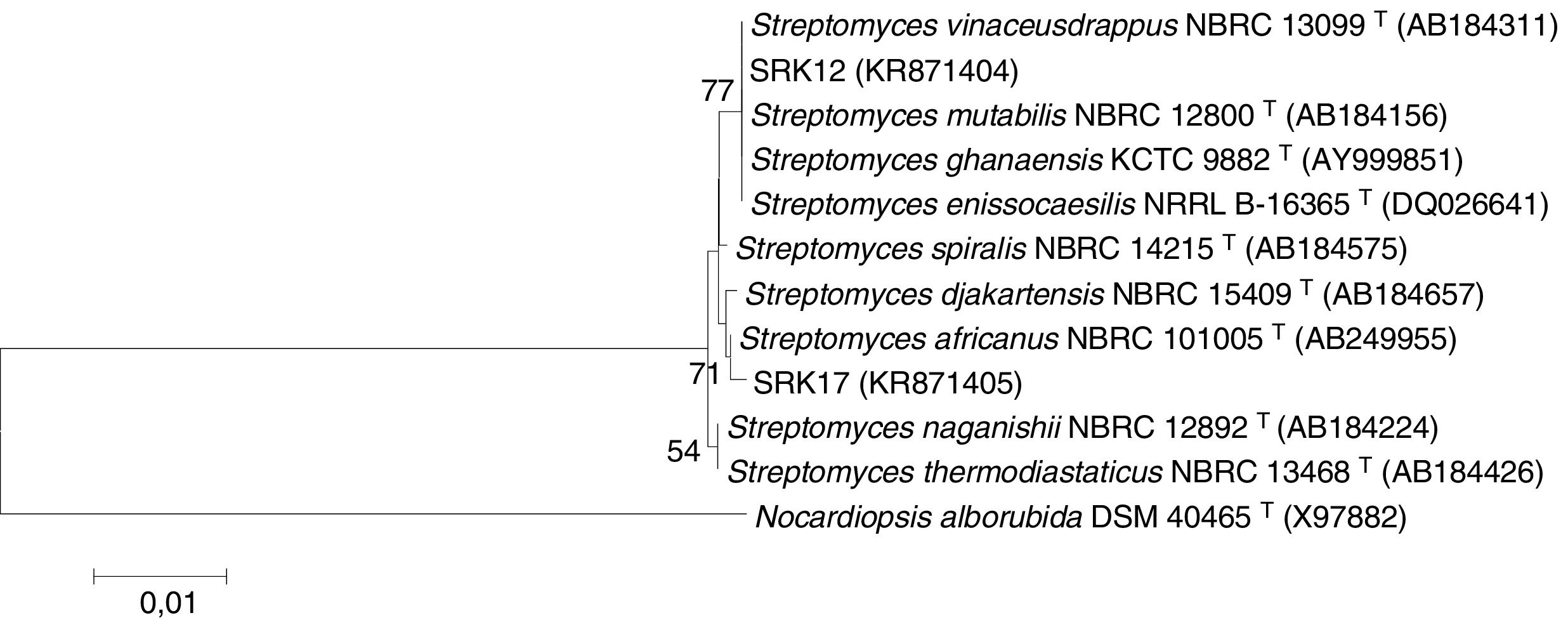
The sequences obtained were compared with 16S rRNA gene sequences of reference strains available at the public Genbank database (www.ncbi.nem.nih.gov) using the BLAST tool. The sequences were aligned using the ClustalW program. The phylogenetic reconstruction (Fig. 1) was done using the neighbor-joining algorithm,29 with bootstrap values calculated from 1000 replicate runs, using MEGA software, version 6.
Phylogeny of SRK12 and SRK17 TBM-resistant strains based on 16S rRNA gene sequences analysis (Tamura-Nei algorithm and neighbor-joining tree). Nocardiopsis alborubida was used as out-group. Bootstrap values (1000 replicate runs) greater than 50% are indicated. GenBank accession numbers are indicated in brackets.
Tribenuron-methyl (methyl 2-[4-methoxy-6-methyl-1,3,5-triazin-2-yl(methyl)carbamoylsulfamoyl]benzoate) was purchased from Sigma–Aldrich (France). Starter cultures were conducted by inoculating 100mL of tryptone-yeast extract broth (5g tryptone, 3g yeast extract per liter of distilled water, pH 7.0–7.2) supplemented with 30mgL−1 of tribenuron-methyl (TBM) with a sporal solution (spore material on 10mL of sterile distilled water). Incubation was done under shaking (150rpm) at 30°C for 48h.22 In order to get a free carbon source inoculum, 100mL of the starter cultures were strongly agitated to break up the Actinobacteria pellets, then centrifugated at 4000×g for 10min. The sediment was washed twice with sterile 0.9% NaCl and suspended in 75mL of sterile physiological water. The resulting suspension is the washed inoculum.22
In order to assess the effect of TBM on Actinobacteria growth, mineral salts ISP9 broth (2.64g (NH4)2SO4, 2.38g KH2PO4, 5.65g K2HPO4·3H2O, 1g MgSO4·7H2O, 1mL of trace salts solution [0.1g FeSO4·7H2O, 0.1g MnCl2·4H2O, 0.1g ZnSO4·7H2O in 100mL of distilled water, pH 7.0] per liter of distilled water, pH 7.0),22 containing 30mgL−1 of TBM, was inoculated with the washed inoculum of each selected strains and incubated at 30°C, 150rpm. Under the same conditions, two other tests were also performed on [ISP9+TBM (30mgL−1)+glucose (1%)] and [ISP9+glucose (1%)]. This last one was used as a control to evaluate the optimum growth of the strains. All assays were done in triplicate. After three weeks of incubation, the cultures were centrifuged (12,000×g/10min/4°C). The biomass was dried at 105°C until constant weight and the filtered supernatants were submitted to HPLC-UV analysis to measure the TBM dissipation. The HPLC equipment consisted of a Spectra Series P200 pump coupled to an ERC 3415α eluent degasser module, a Spectra Series AS100 autosampler, a Spectra System UV6000 LP UV-visible detector (220nm) and a C18 column (Phemenex, Gemini NX; 25cm×4.6mm, 5μm). The mobile phase contained acetonitrile/water (containing 0.1% of H3PO4) 90:10, v/v, at a flow rate of 1mLmin−1. Aliquots (10μL) of culture media were injected and run at room temperature and the different concentrations of TBM were determined relative to the peak area detected in blank control samples made with TBM solutions on the 10–50mgL−1 range.
Statistical analysisEffect of tribenuron-methyl on bacterial growth, was tested following 2-way ANOVA between the tested media and each selected strains, with an Alpha threshold of 0.05, using STATISTICA® software, version 10, StatSoft, France.
ResultsActinobacteria isolationThe isolation strategy was designed with the aim of selecting Granstar®-resistant Actinobacteria with TBM degradation capacity. In total, forty-two (42) Actinobacteria strains were isolated from the different soils. The repartition of isolates according to their soil origin is shown in Table 1. It is noteworthy that the largest number of isolates was obtained from the untreated agricultural soil 2 with 16 isolates. The number of isolates from the other soils, including treated soil 2, was lower (from 4 to 9 isolates).
TBM resistanceAll isolates resisted to the tested doses, until the critical herbicide concentration of 0.3% (v/v), at which some of them were inhibited. At that level, 11 strains were still growing and showed various degrees of resistance to the increased herbicide doses (Table 2). Among this resistant group of 11 strains, 4 isolates were recovered from untreated soils 2 and 3. As expected, more than half of the resistant strains were isolated from treated soils 1, 2 and 3, and the most resistant strains that grew at the highest herbicide concentrations of 0.6, 0.55 and 0.5%, were isolated from treated soils 3, 1 and 2, respectively.
Characterization of the most resistant strainsThe two most TBM-resistant strains, SRK12 and SRK17, were selected for further characterization. Morphological, physiological and phylogeny based on 16S rRNA gene sequences analyses were performed. Both strains developed a non-fragmented substrate mycelium and aerial mycelium with spiral spore chains (Table 3). They also presented some other common characteristics, namely, rounded spores arranged in chains of under twenty spores length; no melanin pigment production and affiliation to the gray color series, as concluded from their spore color on ISP4 agar. However, the strains SRK12 and SRK17 differed from each other in diffusible pigments production on starch-casein agar, where a pinkish brown pigment was produced by SRK12, while, a rusty brown pigment was detected with SRK17. Also, the substrate mycelium of the strain SRK17 presented some sporulation unlike the SRK12.
Morphological characteristics of SRK12 and SRK17 isolates and their close relative strains.
| Strains | Spore chains morphologya | Spore chains lengtha | Spores morphologya | Spore colorb | Production of diffusible pigmentsc | Melanin on ISP6 | Melanin on ISP7 |
|---|---|---|---|---|---|---|---|
| SRK12 | Spirals | <20 | Rounded | Pale gray | Pinkish brown | – | – |
| SRK17 | Spirals | <20 | Rounded | Wooly gray | Rusty brown | – | – |
| S. vinaceusdrappus | Spirals | >50 | Smooth | Red | – | – | – |
| S. mutabilis | Retinaculiaperti | 3–10 | Smooth | White | – | – | – |
| S. ghanaensis | Spirals | nd | Hairy | Green | nd | nd | nd |
| S. enissocaesilis | Spirals | nd | Smooth | Gray | – | Poorly developed | Poorly developed |
| S. africanus | Spirals | >50 | Spiny | Blue | – | – | – |
Analysis of the 16S rRNA gene sequences of strains SRK12 and SRK17 confirmed their affiliation to the Streptomyces genera. They presented high sequence similarity (99.9%) with sequences related to Streptomyces type strains deposited at the public database GenBank. The phylogenetic tree using Nocardiopsis alborubida DSM 40465T sequence (X97882) as out-group (Fig. 1) showed that strain SRK12 grouped in a single cluster with the type strains of Streptomyces vinaceusdrappus NBRC 13099T (AB184311), S. mutabilis NBRC 12800T (AB184156), S. ghanaensis KCTC 9882T (AY999851) and S. enissocaesilis NRRL B-16365T (DQ026641), with a bootstrap support value of 77%. The strain SRK17 was found related to Steptomyces africanus NBRC 101005T (AB249955), with a bootstrap support rate of 71%.
Effect of TBM on bacterial growthAccording to Fig. 2, both strains showed an insignificant growth (p>0.05) when TBM was used as sole carbon source. On the other hand, when glucose was added, both SRK12 and SRK17 strains growth was evident and significant (p<0.05), but it did not reach the highest level obtained on glucose without TBM (p<0.05).
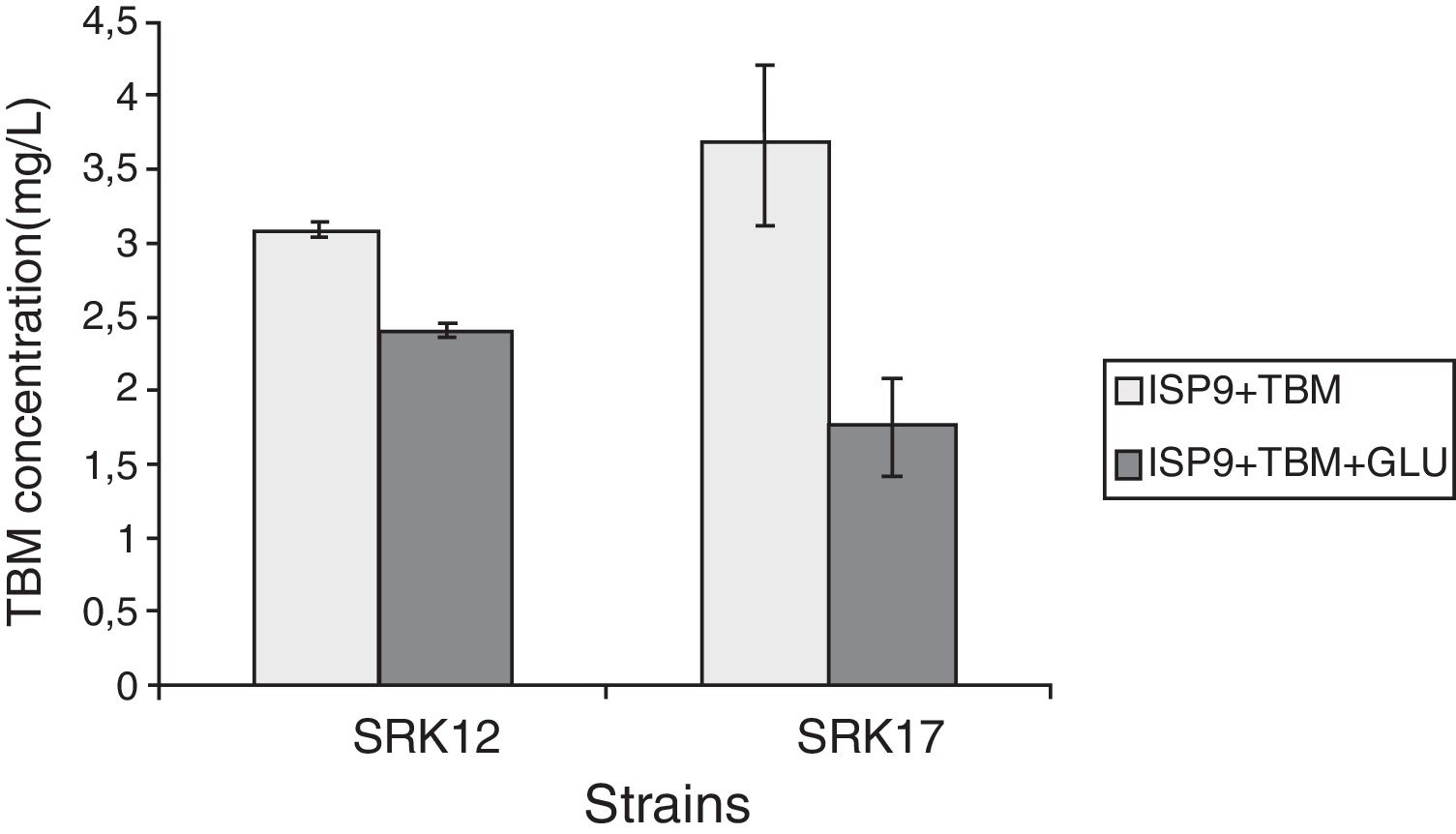
TBM dissipationThe dissipation of TBM varied according to the medium. Fig. 3 shows that, after three weeks of incubation on [ISP9+TBM], the concentration of the studied compound has decreased from 30mgL−1 to 3.09 and 3.66mgL−1, respectively with SRK12 and SRK17 strains. On [ISP9+TBM+glucose], the TBM degradation was improved, the final TBM concentration reaching 2.39 and 1.75mgL−1 respectively.
DiscussionThe low isolate numbers obtained from the three treated soils, suggested that the herbicide treatment reduced the Actinobacteria diversity. This decrease could be explained because of the Granstar® toxicity. Indeed, the presence of TBM, a sulfonylurea molecule, may prevent bacterial growth as suggested by Chèvre (2007)2 and Bottaro et al. (2008).30 These results are in accordance with those of Filimon et al. (2012)31 and Arabet et al. (2014)13 showing different behavior of some bacterial groups, including Actinobacteria species. Sensitive and resistant species were reported after treatment with sulfonylurea herbicides, including some species taking advantage for growth. In our study, the variable number of Actinobacteria isolates according to the different soils suggested that the bacterial communities include sensitive and resistant strains.
According to Arabet et al. (2014),13 the high resistance shown by Actinobacteria isolates could have two possible explanations: either the strains possess a modified acetolactate synthase, making the herbicide inactive on cellular division, or they have the necessary enzymatic luggage to degrade partially or completely the herbicide. The fact that the most resistant strains were isolated from treated soils, suggests that a prior exposure to the herbicide makes bacterial adaptation easier, with even, a possibility to develop a potential for the herbicide transformation. Such a case was observed by Widehem et al. (2002)32 who have isolated from diuron treated soil, a strain of Arthrobacter sp. able to transform the herbicide completely in 24h. Results going in the same direction were reported by Zhao et al. (2015),33 with a TBM-adapted Ochrobactrum sp. strain, which can degrade 55% of the herbicide in 8 days.
Both the SRK12 and SRK17 strains have grown by developing vegetative mycelium and aerial mycelium with spiral spore chains at its extremity. These features supported their assignment to the Streptomyces genera.34,35
According to Fig. 1, the SRK12 strain clustered with four strains of Streptomyces type: S. vinaceudrappus, S. mutabilis, S. ghanaensis and S. enissocaesilis. Knowing that the first three mentioned Streptomyces type strains belong respectively, to red, white and green color series,36–39 the strain SRK12 being from the gray color series, could not be affiliated to them. Although S. enissocaesilis was from the gray color series, it had no diffusible pigments in starch-casein agar,39,40 unlike the strain SRK12. The second isolate SRK17 was closely related to another Streptomyces type-strain named S. africanus, but this one was from the blue color series,39,41 thus it could not be associated to the strain SRK17. Therefore, the discrepancies between the 16S rRNA gene-based affiliation and the color series properties highlighted taxonomic issues as previously indicated by Kämpfer (2006)42 reporting difficulties to differentiate Streptomyces members based on phenotypic features. Such observation suggested that further analyses are required to revisit the Streptomyces genera taxonomy. In our study, the most resistant isolates were then Streptomyces strains. Works conducted by Filimon et al. (2012)31 and Arabet et al. (2014)13 also showed that Streptomyces species were present among sulfonylurea resistant microorganisms.
Our result clearly showed that both strains were unable to use TBM as sole carbon source, hence they did not have the necessary enzymes to degrade it. Wang et al. (2012)43 as well as Zhang et al. (2013)44 also noticed that Serratia sp. and Pseudomonas sp. strains, respectively, were unable to grow when inoculated on mineral salts broth supplemented with TBM. The non-significant development of both SRK12 and SRK17 strains in presence of TBM can be explained by the presence of residual carbon sources, from starter cultures, that may remain confined in the Actinobacteria pellets biomass, in spite of the use of a twice washed inoculum. On this medium composition, the degradation of TBM was considerable leading to low concentrations of 3.09 and 3.66mgL−1 respectively with SRK12 and SRK17 strains (Fig. 3). Since the tested strains were not able to grow on TBM, as sole carbon source (Fig. 2), the sulfonylurea molecule has probably been degraded by a chemical pathway, without a bacterial involvement. Several studies8,12,43,45 reported that acidohydrolysis was the primary dissipation mechanism of TBM in the soil environment and aqueous solutions. This process was mainly influenced by variation of pH value, i.e., the degradation occurred more rapidly at lower pH than at higher pH.3 Wang et al. (2012)43 mentioned that at pH 7.3, TBM was not degraded at all. The same observation was noted by Zanardini et al. (2002)46 for two sulfonylurea compounds; chlorsulfuron and metsulfuron methyl, at 7.0–7.2. In our experiment, the final pH was 6.72 with both strains, which is close to pH value of 6.8 that Martin (2000)12 considered as a critical point for sulfonylurea herbicides degradation. Thus this acidity level was probably the trigger factor for the dissipation of TBM by acidohydrolysis mechanism. It is interesting to notice that, as the TBM decreased in the first hours of incubation, two compounds were detected at HPLC, with peaks areas increasing over time (data not shown). These products were probably derived from the cleavage of the sulfonylurea bridge of TBM which is the most common and the major initial chemical reaction for sulfonylurea breakdown, especially under acidic conditions.3,8,12,44,45 From these results, two conclusions can be drawn; both strains were also unable to use TBM degradation products as a carbon source; and unlike TBM, these compounds were not degraded either chemically or biologically.
The addition of another carbon source (glucose) with TBM has allowed the growth of both Actinobacteria strains (Fig. 2), with even an improvement of TBM dissipation rate, leading to concentrations of 2.39 and 1.75mgL−1 respectively with SRK12 and SRK17 strains (Fig. 3). This can be explained by the decrease of pH values to 4.38 and 4.23 respectively, that were likely obtained by the accumulation of organic acids produced from glucose fermentation, as suggested by Zanardini et al. (2002).46 These acidic conditions that may enhance TBM transformation would then be directly related to bacterial activity on glucose catalysis. Accordingly, we concur with Wang et al. (2012)43 in suggesting that TBM degradation could be a co-metabolic process involving bacterial mediated acidohydrolysis of TBM.
The significant differences noticed between strains growth on glucose, with and without TBM (Fig. 2), was not due to TBM since it was almost entirely dissipated, but it can be explained by the presence of one or both new detected compounds from TBM degradation, which incontestably seems to hamper the development of both strains, even when glucose was added. This observation confirmed the toxic effect of TBM transformation on tested actinobacteria, but it was not in accordance with that found by Wang et al. (2012)43 and Zhang et al. (2013)44 who reported no evident effect of TBM on Serratia sp. and Pseudomonas sp. cell yield, respectively, when inoculated on glucose with and without TBM. This insinuated different degrees of TBM-induced transformation and different bacterial behavior.
It is known that pesticides by-products may have a more pronounced toxicity than the precursor molecule. That's what appears to be the case here, where the probable cleavage of the TBM sulfonylurea bridge has led to the appearance of two compounds significantly interfering with bacterial growth. It is most likely be sulfonamide and triazine amine which are considered to be stable and highly persistent in soils.3,4,30,45 The accumulation of those two metabolites at high concentrations could lead to the inhibition of the most resistant actinobacteria.
ConclusionThe isolation strategy adopted in this study confirmed the negative effect of Granstar® herbicide on Actinobacteria. Resistant strains to high levels of the TBM were obtained. The two most resistant strains SRK12 and SRK17 were assigned to the Streptomyces genera on the basis of morphological characteristics and 16S rRNA gene sequences. However, discrepancies were observed between 16S rRNA gene-based affiliation and color properties highlighting Streptomyces taxonomic issues, which should require in depth taxonomic studies of the Streptomyces genera.
The first reaction of the degradation of TBM can be triggered under acidic conditions by a co-metabolic process involving bacterial-mediated acidohydrolysis, which would finally result in the partial breakdown of the TBM molecule. This process would be a promising application in bioremediation of neutral and alkaline TBM-polluted soils. The products of this chemical degradation seem to have a harmful effect on the actinobacteria growth. For a better understanding of the whole process, further tests are needed, including the identification of the resulting metabolites and the study of their destiny by soil microbial consortium.
Conflicts of interestThe authors declare that they have no conflicts of interest.
This work was supported by the Université Larbi BenMhidi, Oum El Bouaghi (Algeria), the “Equipe Environnement et Microbiologie” (EEM), MELODY group, UMR IPREM 5254, Université de Pau et des Pays de l’Adour (France), and the Laboratoire de Génie Microbiologique et Applications, Université Frères Mentouri, Constantine 1 (Algeria).