Unraveling the microbial diversity and its complexity in petroleum reservoir environments has been a challenge throughout the years. Despite the techniques developed in order to improve methodologies involving DNA extraction from crude oil, microbial enrichments using different culture conditions can be applied as a way to increase the recovery of DNA from environments with low cellular density for further microbiological analyses. This work aimed at the evaluation of different matrices (arenite, shale and polyurethane foam) as support materials for microbial growth and biofilm formation in enrichments using a biodegraded petroleum sample as inoculum in sulfate reducing condition. Subsequent microbial diversity characterization was carried out using Scanning Electronic Microscopy (SEM), Denaturing Gradient Gel Electrophoresis (DGGE) and 16S rRNA gene libraries in order to compare the microbial biomass yield, DNA recovery efficiency and diversity among the enrichments. The DNA from microbial communities in petroleum enrichments was purified according to a protocol established in this work and used for 16S rRNA amplification with bacterial generic primers. The PCR products were cloned, and positive clones were screened by Amplified Ribosomal DNA Restriction Analysis (ARDRA). Sequencing and phylogenetic analyses revealed that the bacterial community was mostly represented by members of the genera Petrotoga, Bacillus, Pseudomonas, Geobacillus and Rahnella. The use of different support materials in the enrichments yielded an increase in microbial biomass and biofilm formation, indicating that these materials may be employed for efficient biomass recovery from petroleum reservoir samples. Nonetheless, the most diverse microbiota were recovered from the biodegraded petroleum sample using polyurethane foam cubes as support material.
Crude oil biodegradation in petroleum reservoirs affects the world production of fuels, making the recovery and refining processes more expensive. For many years, the prevalent occurrence of biodegradation in petroleum wells has been attributed to the aerobic bacterial degradation of hydrocarbons, which can be stimulated by oxygen carried by the infiltration of meteoric waters in the reservoir.1 However, there is strong evidence for the widespread occurrence of obligate anaerobes in subsurface petroleum systems,2–4 and the flushing of meteoric water does not indicate that highly reactive oxygen survives transportation to deep reservoirs, since even small concentrations of organic compounds can remove oxygen from an aquifer.
In recent work, researchers have suggested that biodegradation processes can occur at the oil–water transition zone, in which microbial life should be possible within water droplets containing active microbial communities.5 Data gathered from several studies indicate that oil biodegradation in deep subsurface petroleum reservoirs occurs through anaerobic microbial metabolism rather than aerobic mechanisms, resulting in a decrease of light hydrocarbons and an increase of oil density, acidity, viscosity and sulfur content.6–8 In addition, viable anaerobic hydrocarbon degradation processes have recently been established for both saturated and aromatic hydrocarbons.9–11 Studies aiming to evaluate intermediate metabolites, characteristic of anaerobic hydrocarbon degradation, have been carried out and allowed the identification of compounds such as reduced 2-naphthoic acids,12 2-methylnaphthalene, tetralin, as well as naphthoic acids in petroleum reservoirs.13
Sulfate reduction and methanogenesis are the most likely processes responsible for in-reservoir hydrocarbon oxidation.14 Oil degradation linked to sulfate reduction would explain the consistent hydrocarbon compositional patterns seen in many degraded oils worldwide. Sulfate arises from geological sources, such as evaporitic sediments and limestone, or from the injection of seawater for pressure stabilization, and may lead to significant oil degradation and increased residual-oil sulfur content.15 Souring in oilfield systems is most commonly due to the action of sulfate-reducing prokaryotes, a diverse group of anaerobic microorganisms that respire sulfate and produce sulfide (the key souring agent) while oxidizing diverse electron donors.8
In this sense, efforts have been made by several researchers in order to recover and characterize the anaerobic microbial community inhabiting the deep petroleum biosphere.2,16,17 The study of genomes of uncultivated microbes have become possible through metagenomics, a cultivation-independent approach that allows to explore the metabolic potential of the unseen biodiversity by cloning large DNA fragments directly isolated from the environment.18 With the use of the metagenomic approach, bacteria capable of degrading petroleum hydrocarbons, including anaerobes, have been more deeply investigated and their metabolic routes unraveled.19,20 However, the extremely low amount of DNA obtained from samples derived from petroleum reservoirs using direct nucleic acid extraction procedures is often a restraint when the phylogenetic and/or metabolic diversity of microbial communities are investigated,21,22 because of the low cellular density and activity found in such hostile environment. Microbial enrichments using different culture conditions, simulating the chemical and physical parameters of natural environments, can be applied to overcome this limitation and increase the recovery of DNA from environments with low cellular density.23 Although cultivation under laboratory conditions can diminish the biodiversity recovered, this technique allows the selection of microorganisms that have some function of particular interest, such as enzymatic activity or biodegradation ability.24
This work aimed to evaluate the efficiency of different matrices, used as physical supports, in recovering anaerobic bacterial diversity from a biodegraded oil sample derived from a petroleum reservoir in Campos Basin (Brazil). The matrices were used in order to evaluate their effect in the increase of biomass, as well as a support for biofilm formation. The relative abundance and diversity of the anaerobic microbiota recovered from the enrichments were compared by using microscopic and molecular analysis (DGGE and 16S rRNA libraries).
Material and methodsSamplingPetroleum samples were obtained in July 2005 from five production wells at the Pampo Platform, Campos Basin (Macaé, RJ, Brazil), with logistic support from CENPES/Petrobras, as described in details by Vasconcellos et al.22 Samples were collected in triplicate using 500mL sterilized Schott bottles, which were completely filled with the samples in order to avoid oxygen influx. Samples were kept on ice during transportation to the laboratory and stored at room temperature for subsequent anaerobic bacterial enrichment assays.
Anaerobic enrichmentsThe biodegraded petroleum sample (P2) used in this work as inoculum (10% v/v) for the anaerobic enrichments was collected from oil reservoir 2 in Campos Basin, RJ, Brazil.22 This well was characterized as highly biodegraded, level 5–6, according to Peters and Moldowan,25 with average temperature 71°C and approximately 2000m deep. The petroleum sample was homogenized in water bath at 50°C. The enrichments were settled in Schott bottles (1L) containing 500mL of Zinder medium26 supplemented with organic substrates (sodium acetate, sodium formate, sodium lactate, yeast extract, methanol) to stimulate the growth of sulfate reducing bacteria, according to methods described by Dubourguier et al.27 and Silva et al.28. The cysteine–HCl solution (2mM) was added to the enrichments (1%, v/v) as final electron acceptor.
Three different matrices were independently applied as physical supports to allow bacterial biofilm formation and increase biomass recovery under sulfate reducing condition: (1) polyurethane foam cubes (PF) (1cm2), (2) slices of shale (S), and (3) arenite (A). Bacterial enrichment using P2 as inoculum but without any matrix was settled as control (WS). Each condition was evaluated in triplicate.
The polyurethane foam cubes were submitted to UV sterilization for 20min followed by immersion in autoclaved Zinder medium, under anaerobic condition (N2 flow). Approximately 80 cubes were added to each flask of microbial enrichment under condition (1). Samples of shale and arenite, which are natural components of oil reservoirs, were kindly donated by Dr. Eugenio V. dos Santos Neto (CENPES/Petrobras). Shale was sliced in small pieces, while arenite was grated in powder. Both materials were autoclaved twice in Schott bottles (250mL) containing 200g of slices or powder. After sterilization, 10g of each matrix were inoculated in the corresponding microbial enrichments.
All anaerobic enrichments were incubated at 55°C, during 60 days, in a rotary shaker at 100rpm.
Microscopic analysisScanning electronic microscopy (SEM) was used to evaluate the abundance and diversity of cell morphology of all bacterial enrichments under study. The analyses were developed with a Zeiss microscope model LEO 982, at Embrapa/CNPMA, using protocols described by Melo et al.29.
DNA extractionThe DNA extraction from the bacterial enrichments was carried out using a protocol based on Groβkopf et al.30 and Neria-Gonzáles et al.,31 with adaptations to petroleum samples. Firstly, 50mL of a sterilized Tween 80 solution (10%) (Sigma–Aldrich) were added into enrichments in order to promote homogenization of oil/water phases, as well as improving the recovery of cells adhered on the physical supports (foams, shale and arenite). Total enrichment volume was distributed in sterile tubes (50mL) and centrifuged at 10,000rpm, for 20min, 4°C. Supernatants were discarded and cells were transferred to microtubes (2mL). Afterwards, microbial pellets retrieved from the enrichments (3×500mL) were suspended in 600μL PBS buffer, homogenized by vortex and lysozyme was added at a final concentration of 17mg/mL. After incubation at 37°C for 2h, proteinase K and SDS were added (final concentration of 0.7mg/mL and 2%, respectively) and the solution was incubated at 60°C for 90min. The microtubes were submitted to three freeze–thaw cycles (2min in liquid nitrogen followed by 2min at 65°C). Glass beads were added to the tubes and manual agitation was performed for 1min. The solution was extracted once with equal volume of saturated phenol (pH 8.0) and once with equal volume of chloroform:isoamyl alcohol (24:1). For DNA precipitation, 5M NaCl (10%) and 2 volumes of cold ethanol were added to the solution. The pellet was washed once with ethanol 70%, dried and suspended in Milli-Q water. The yield and integrity of the DNA obtained were confirmed through NanoVue Plus™ Spectrophotometer (GE Healthcare) and electrophoresis in 1% agarose gel stained with ethidium bromide and documented using a UVP BioImaging System GDS-8000 (UVP, Upland, CA, USA).
Construction of 16S rRNA gene libraries, ARDRA and phylogenetic analysesFor the construction of the 16S rRNA gene libraries, amplification was performed from total community DNA, obtained from each enrichment, by using the bacterial primer set 27f and 1100r.32 Only one library was assembled for each enrichment. Fifty microliter-reaction mixtures were made contained 50ng of total DNA, 2 U of Taq DNA polymerase (Invitrogen), 0.2mM of dNTP mix and 0.4μM of each primer, in 1X Taq buffer. The PCR amplifications were performed using 10 cycles of 1min at 94°C, 30s at 60°C, decreasing 0.5°C each cycle, and 3min at 72°C, followed by another 10 cycles of 1min at 94°C, 30s at 56°C and 3min at 72°C. Amplicons were pooled from five reactions (∼500ng), purified using GFX™ PCR-DNA and gel band purification kit (GE Healthcare) and cloned using the pGEM-T cloning vector kit, according to the manufacturer's instructions (Promega, Madison, Wisc.). Insert-containing clones were submitted to ARDRA by digestion of M13 amplicons with the enzymes Hae III, Hha I and Msp I, independently, at 37°C for 2.5h. Clones representing distinct ribotypes were selected for DNA sequencing and phylogenetic affiliation.
The 16S rRNA gene sequences were determined by direct amplification of the inserts from overnight grown clone cultures with M13 forward and reverse primers and sequencing with the DYEnamic ET Dye Terminator Cycle Sequencing Kit for the automated MegaBace 500 system (GE Healthcare) using the primers 10f, 1100r, 765f and 782r,32 according to the manufacturer's recommendations. Partial 16S rRNA gene sequences obtained from clones were assembled in a contiguous sequence using the phred/Phrap/CONSED program.33,34 Phylogenetic affiliation was achieved as described previously by Vasconcellos et al.22.
The nucleotide sequences determined in this study were deposited at the Genbank database under the accession numbers: GenBank ID: JN998802 to JN998890.
DGGE analysesThe PCR targeting 16S rDNA for the DGGE analyses was performed using the universal primers 968f (attached to a 40-nucleotide GC-rich sequence) and 1401r,35 which are homologous to the conserved bacterial 16S rDNA regions. The PCR amplifications were performed in 50μL reactions containing 50ng of total community DNA recovered from the microbial enrichments, 5μL of 10× Tris–HCl reaction buffer, 1.5mM MgCl2, 0.4μM primers 968f and 1401r, 0.2mM dNTP mix and 2 U Taq DNA Polymerase (Invitrogen, Grand Island, N.Y., USA). The PCR amplifications were performed using an initial denaturation step of 5min at 94°C, 10 cycles of 1min at 94°C, 30s at 58°C, decreasing 1°C each cycle, and 2min at 72°C, followed by another 25 cycles of 1min at 94°C, 30 s at 53°C and 2min at 72°C. The amplicons were first checked on 1.2% agarose gels prior to the DGGE analyses.
The DGGE analyses were carried out in the D-Code Universal Mutation Detection System (Bio-Rad, USA) using a linear denaturing gradient of urea and formamide ranging from 50% to 70% (100% denaturant corresponding to 7M urea and 40% (v/v) deionized formamide). Gels (6% polyacrylamide) containing 6μL of PCR products for each sample, in triplicate, were run at 50V and 60°C for 14h in 0.5× TAE buffer. Gels were stained with SYBR Green 1× solution and documented under UV light.
ResultsMicroscopic analyses of bacterial enrichmentsA dense cellular biomass (up to 108cells/mL) was observed when using polyurethane foams as matrices in the anaerobic enrichments after 60 days of incubation. The enrichments without physical supports exhibited low medium turbidity (104cells/mL) when compared to the others in which matrices were employed.
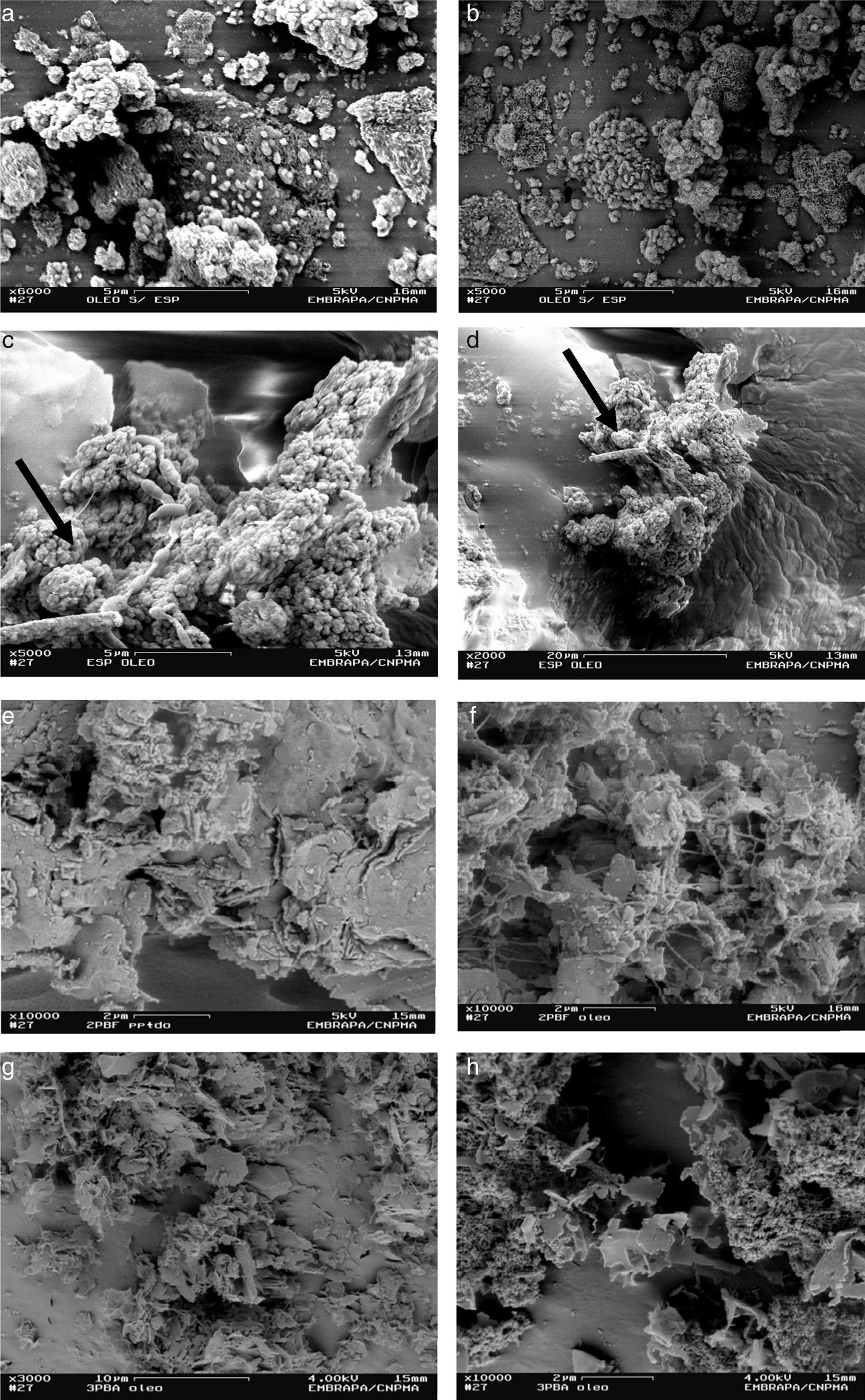
The SEM analyses demonstrated that bacterial enrichments without physical supports yielded low abundance of cells and no biofilm formation (Fig. 1a and b). Actually, in this condition cells were shown to be sparsely distributed. On the other hand, a dense biomass yield and biofilm formation could be observed around (shale) or inside the porous (arenite and polyurethane foam) of the other matrices (Fig. 1c–h).
A predominance of coco rods was detected in the enrichments without physical supports (Fig. 1a and b), whereas straight rods forming polymeric structures (EPS) were observed when using polyurethane foams (Fig. 1c and d). The use of arenite and shale as supports allowed an intense biofilm formation involved by EPS, and thus the determination of the microbial morphology was not possible (Fig. 1e–h).
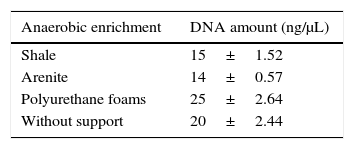
DNA extraction and DGGE analysesAfter 60 days of incubation, the recovery of community DNA was possible for all the petroleum-based anaerobic enrichments performed in this study. It is worth to mention that different controls were set up, using only the matrices and sterilized medium. The controls did not show any turbidity, demonstrating that no contamination occurred. Although the enrichment without physical support exhibited lower turbidity of cells, the enrichments containing shale and arenite as support showed lower DNA recovery when compared to the enrichments without support and to the one containing polyurethane foams (Table 1). This was probably due to the strong adsorption of bacterial cells to the pieces of shale and arenite granules, making it difficult the recovery of all the biomass developed in these enrichments.
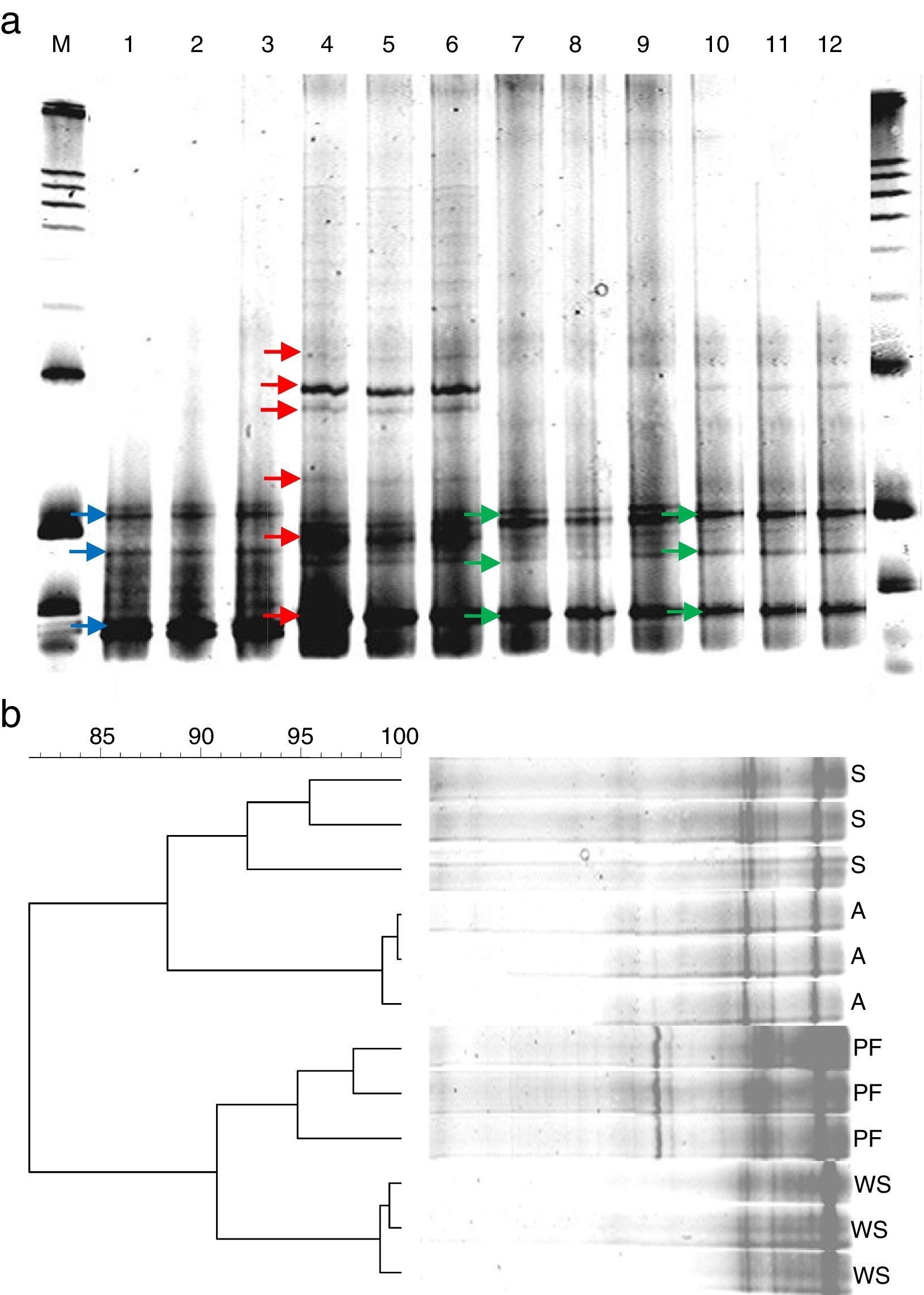
The DGGE analyses revealed distinct band profiles between samples originated from bacterial enrichments with and without physical supports, reflecting differences in the bacterial community composition (Fig. 2a). The type of physical support used in the bacterial enrichments also resulted in differences, in particular dominant populations reflected by the DGGE bands. These differences were observed in terms of number and intensity of bands. Most bands observed in the profiles corresponding to the enrichment without any physical support were not observed in the enrichments with support, indicating that the bacteria related to those bands were not favored under the conditions settled for the other enrichments (Fig. 2a, blue arrows in lanes 1–3). On the other hand, profiles derived from the enrichments containing supports for an increase of biomass revealed specific bands not observed in the enrichment without any physical matrix (Fig. 2a, red and green arrows in lanes 4–12). Enrichments with arenite and shale as matrices were more similar to each other when compared to the enrichment with polyurethane foams (Fig. 2b). In addition, band patterns corresponding to the arenite and shale enrichments showed the lowest relative richness (number of bands); whereas, the enrichments with polyurethane foam showed the highest complex band patterns (Fig. 2a, lanes 4–6).
16S rRNA gene libraries, ARDRA and phylogenetic analysisThe bacterial diversity of the four different enrichment cultures evaluated in this work was determined by analysis of 16S rRNA gene clone libraries.
A total of 95 clones from the WS (without support) enrichment, 46 clones from the PF (polyurethane foams) enrichment, 63 clones from the S (shale) enrichment and 50 clones from the A (arenite) enrichment were screened by ARDRA aiming to select different ribotypes for subsequent sequencing and phylogenetic analysis. Combined data from ARDRA and sequencing analyses allowed to unravel the bacterial diversity recovered in the petroleum enrichments (Fig. 3). Clones were related to sequences available at the Genbank and RDP (Ribosomal Database Project) public database.
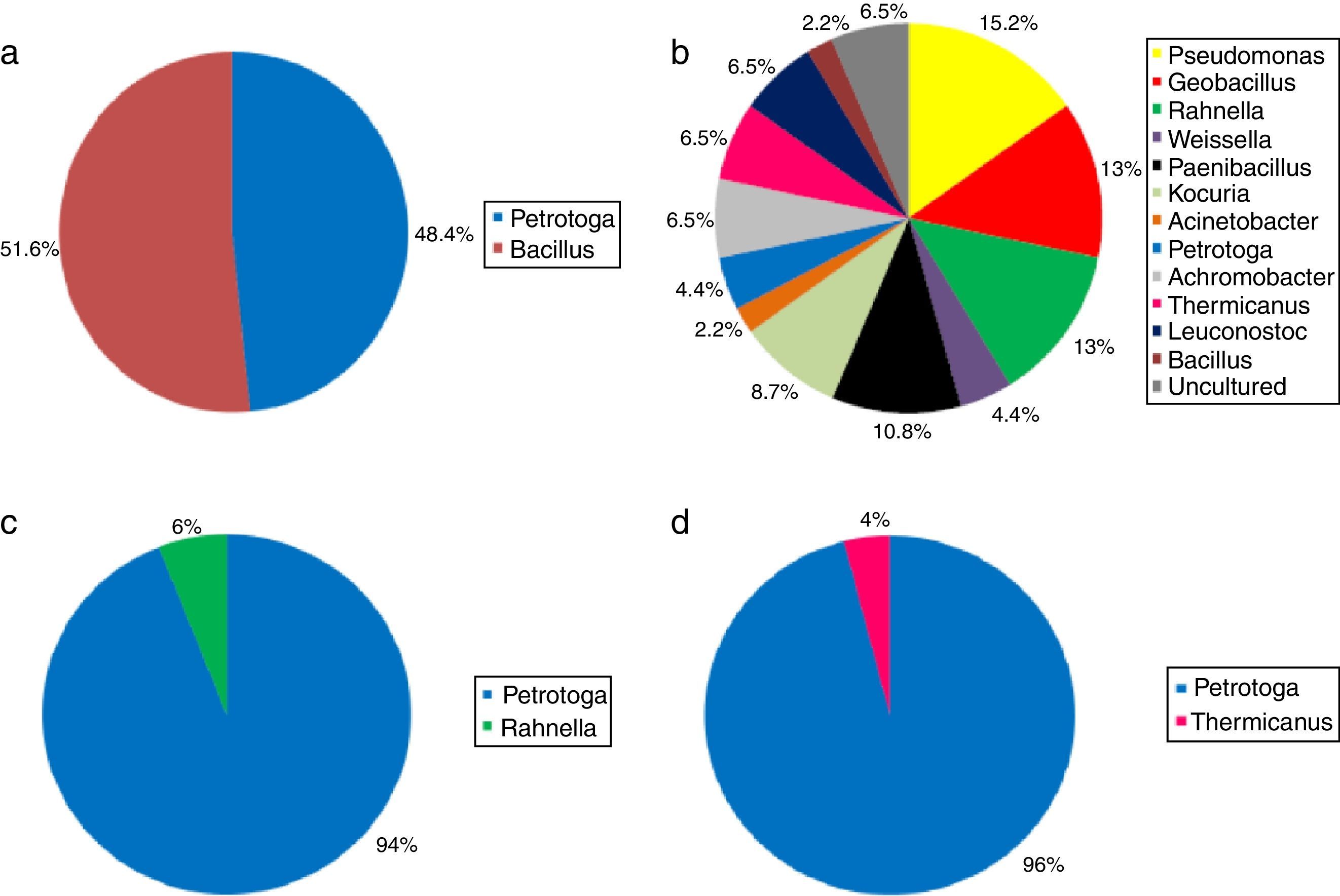
Occurrence of bacterial genera in the petroleum enrichments: (a) Enrichment without support material (WS) (95 clones, 7 ribotypes); (b) enrichment with polyurethane foams (PF) as support material (46 clones, 15 ribotypes); (c) enrichment with shale (S) as support material (63 clones, 3 ribotypes); and (d) enrichment with arenite (A) as support material (50 clones, 3 ribotypes).
The ARDRA analysis of the 95 clones from the WS enrichment showed seven distinct restriction profiles. Clones were affiliated to the genera Petrotoga (48.4%) (Phylum Thermotogae) and Bacillus (51.6%) (Phylum Firmicutes) (Fig. 3a).
Fifteen distinct ribotypes were detected in the PF enrichment. Sequencing of clones representing such ribotypes revealed a more diversified microbiota, which included the genera Rahnella (13%), Pseudomonas (15.2%), Achromobacter (6.5%) and Acinetobacter (2.2%) (Phylum Proteobacteria), Geobacillus (13%), Paenibacillus (10.8%), Thermicanus (6.5%), Weissella (4.4%), Leuconostoc (6.5%) and Bacillus (2.2%) (Phylum Firmicutes), Petrotoga (4.4%) (Phylum Thermotogae) and Kocuria (8.7%) (Phylum Actinobacteria). Some clones (6.5%) were related to sequences from uncultured bacteria, and thus considered unaffiliated (Figure 3b). The ARDRA screening of the 63 clones from the S enrichment yielded three distinct ribotypes. Analyses of the clone sequences revealed that the bacterial community was composed basically by the phyla Thermotogae, represented by the genus Petrotoga (94%), and Proteobacteria, represented by the genus Rahnella (6%) (Fig. 3c).
Finally, ARDRA screening also revealed three distinct ribotypes from 50 clones recovered from the A enrichment. Similarly to the S enrichment, the experiments using arenite as physical support revealed the massive predominance of the genus Petrotoga (Phylum Thermotogae) (96%). Besides Thermotogae, the Phylum Firmicutes was also identified in this microbiota, represented by clones related to the genus Thermicanus (4%) (Fig. 3d).
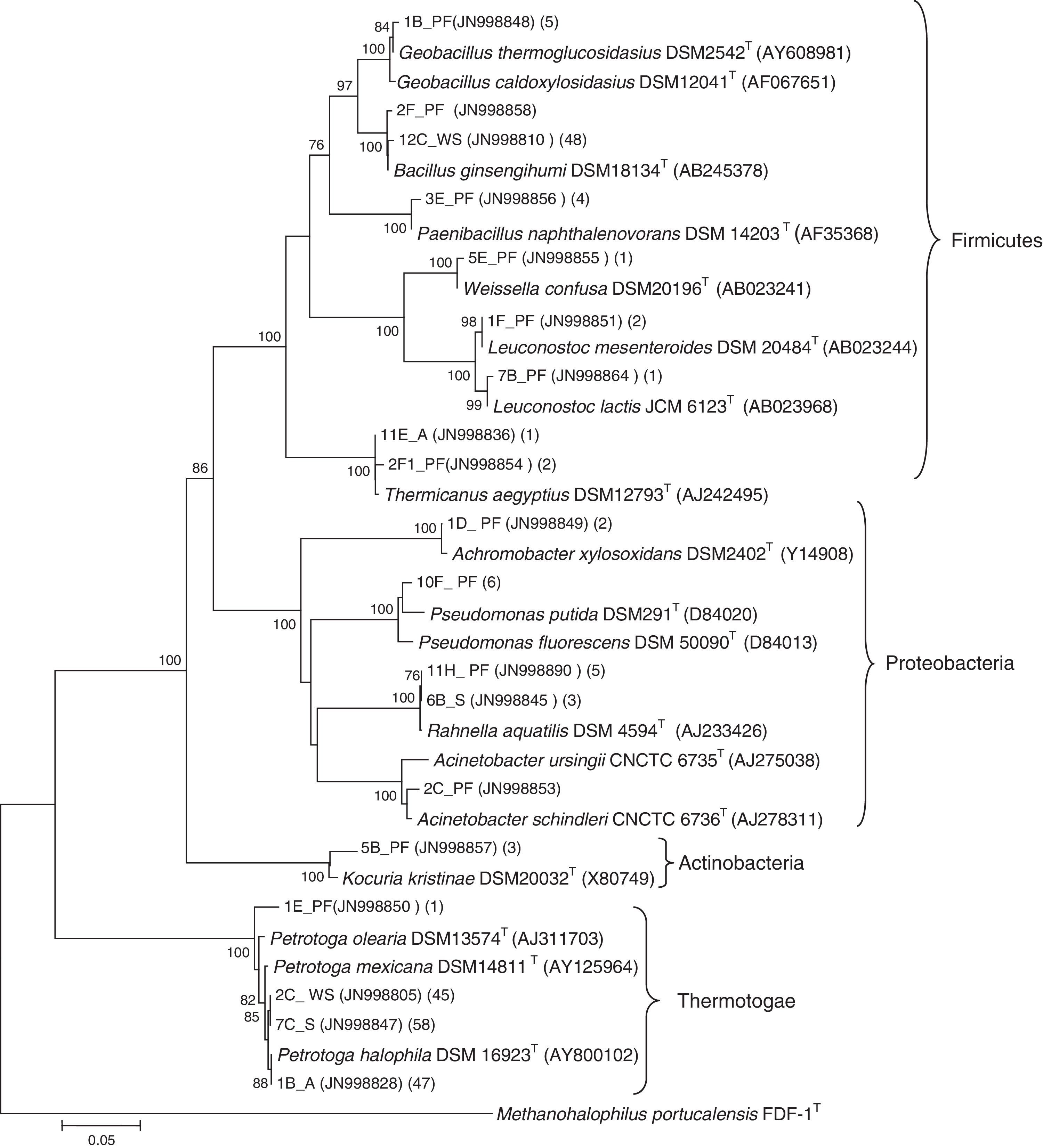
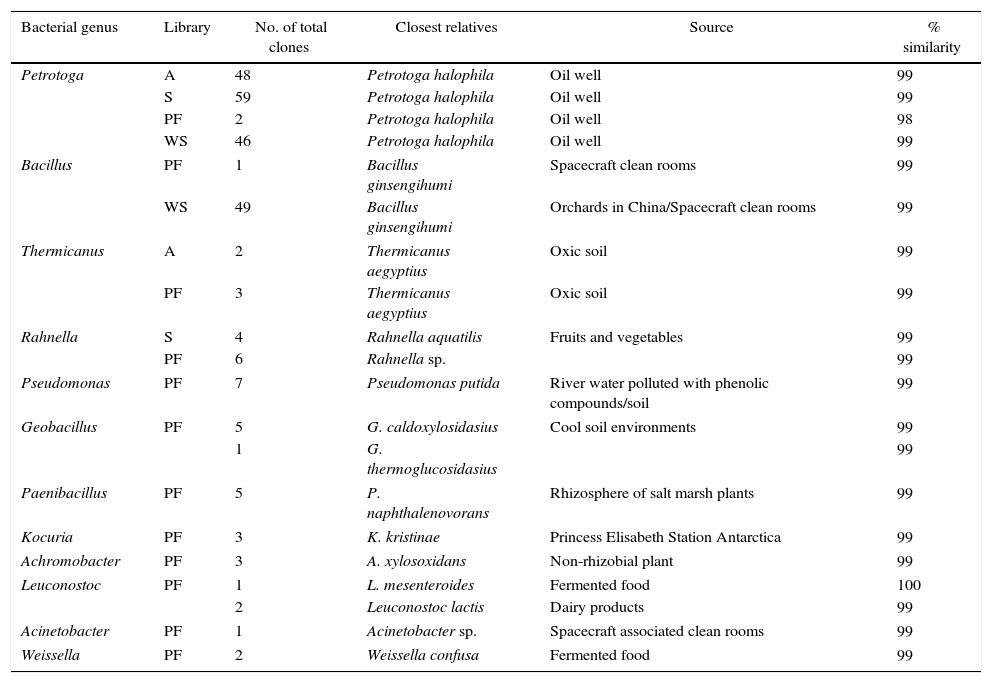
Phylogenetic analysis allowed the identification of many bacterial members at the specie level (Fig. 4). The majority of the clones related to Petrotoga present in the libraries from the A, S, PF and WS enrichments were related to three Petrotoga species, being Petrotoga halophila the closest one. These clone sequences exhibited high sequence similarity (99%) with Petrotoga strains isolated from oil reservoirs (Table 2). Actually, the majority of the analyzed clones in all libraries were affiliated with the genus Petrotoga, amounting to 155 clones, 48 from A, 59 from S, 46 from WS and 2 from PF enrichment. Clones related to Bacillus present in the libraries from the WS (49 clones) and PF (1 clone) enrichments were related to Bacillus ginsengihumi, showing 99% sequence similarity (Table 2). Ten clones present in the S and PF libraries were related to Rahnella aquatilis and Rahnella sp. (sequence similarity between 80% and 99%). Five clones were closely related to the species Thermicanus aegyptius, two from the A library and three from the PF library, showing 99% of sequence similarity.
Phylogenetic analysis based on partial bacterial 16S rRNA sequences of clones from diverse enrichment samples and related species. Bootstrap values greater than 70% are listed. GenBank accession numbers are listed after species names. Numbers in brackets correspond to additional clones presenting. 97% sequence similarity with the clones represented in the tree. Letters PF, WS, A and S correspond to the sample libraries. Methanohalophilus portucalensis was used as outgroup.
Bacterial diversity of anaerobic enrichments from a biodegraded petroleum sample from Campos basin revealed by culture-independent methods.
| Bacterial genus | Library | No. of total clones | Closest relatives | Source | % similarity |
|---|---|---|---|---|---|
| Petrotoga | A | 48 | Petrotoga halophila | Oil well | 99 |
| S | 59 | Petrotoga halophila | Oil well | 99 | |
| PF | 2 | Petrotoga halophila | Oil well | 98 | |
| WS | 46 | Petrotoga halophila | Oil well | 99 | |
| Bacillus | PF | 1 | Bacillus ginsengihumi | Spacecraft clean rooms | 99 |
| WS | 49 | Bacillus ginsengihumi | Orchards in China/Spacecraft clean rooms | 99 | |
| Thermicanus | A | 2 | Thermicanus aegyptius | Oxic soil | 99 |
| PF | 3 | Thermicanus aegyptius | Oxic soil | 99 | |
| Rahnella | S | 4 | Rahnella aquatilis | Fruits and vegetables | 99 |
| PF | 6 | Rahnella sp. | 99 | ||
| Pseudomonas | PF | 7 | Pseudomonas putida | River water polluted with phenolic compounds/soil | 99 |
| Geobacillus | PF | 5 | G. caldoxylosidasius | Cool soil environments | 99 |
| 1 | G. thermoglucosidasius | 99 | |||
| Paenibacillus | PF | 5 | P. naphthalenovorans | Rhizosphere of salt marsh plants | 99 |
| Kocuria | PF | 3 | K. kristinae | Princess Elisabeth Station Antarctica | 99 |
| Achromobacter | PF | 3 | A. xylosoxidans | Non-rhizobial plant | 99 |
| Leuconostoc | PF | 1 | L. mesenteroides | Fermented food | 100 |
| 2 | Leuconostoc lactis | Dairy products | 99 | ||
| Acinetobacter | PF | 1 | Acinetobacter sp. | Spacecraft associated clean rooms | 99 |
| Weissella | PF | 2 | Weissella confusa | Fermented food | 99 |
Other bacterial species were identified only from the PF library (Fig. 4). One clone related to Acinetobacter ursingii was found, showing 99% sequence similarity. Three clones were related to Leuconostoc mesenteroides and two clones to Weissella confusa, both species isolated from fermented food (Table 2), with 99% sequence similarity. Six clones were clustered with high bootstrap value with the type strains of Geobacillus thermoglucosidasius and Geobacillus caldoxylosidasius, showing 99% sequence similarity. In this case, it was not possible to define the identification of the clones at the species level, since they were recovered in a tight cluster with both species. Five clones were closely related to Paenibacillus naphthalenovorans (99% sequence similarity). In addition, four clones were clustered with the actinobacterium Kocuria kristinae (100% bootstrap value), three clones with Achromobacter xylosoxidans (99% sequence similarity; 100% bootstrap value) and, finally, seven clones with the type strain of Pseudomonas putida.
DiscussionIn this study, three different types of matrices (polyurethane foam, shale and arenite) were evaluated as supports for biomass immobilization and increase in anaerobic enrichments from a biodegraded petroleum sample (Campos Basin). Shale, in particular, constitutes nearly 70% of the rocks present in a sedimentary basin. Geochemical analyses showed that almost all hydrocarbons of the petroleum from Campos Basin are from shale, belonging to the Lower Cretaceous Lagoa Feia Formation.36 In this sense, this material is probably of great relevance in providing a substrate for the microbial growth in petroleum reservoirs. The use of shale in bacterial enrichment from the petroleum sample allowed the cells to grow around the shale slices generating a type of biofilm. In fact, cell aggregates, as well as the EPS structure responsible for the maintenance of cell cohesion, were observed for the enrichments employing the two other physical supports, arenite and polyurethane foams, but not for the WS (without support) enrichment. These results confirm the usefulness of these types of physical supports to enable biofilm formation and increase the microbial biomass from low cellular abundance samples, corroborating previous findings.28,37 However, in terms of DNA recovery, polyurethane foams were the most efficient material.
It is known in the literature that bacteria can survive in associations, named as biofilms, producing dense biomass and polymeric substances able to keep them together as a unit.38 In petroleum reservoirs the presence of microbial biofilms are mostly associated with corrosion process.39 In case of specific groups as sulfate reducing bacteria, for instance, it is generally accepted that souring microbiota can form mixed community biofilms on the reservoir mineral matrix.8
Immobilization often simulates what occurs naturally when cells grow on surfaces or within natural structures. Numerous biotechnological processes are incremented by the use of microbial immobilization techniques. These techniques can be divided into four types, based on the physical characteristics of the supports employed: (1) attachment or adsorption on solid carrier surfaces, (2) entrapment within a porous matrix, (3) self-aggregation by flocculation (natural or induced) and (4) cell containment barriers.40 However, the choice of a material as the ideal physical support can be determinant in the selection of a microbial community,28 which could be observed also in this work. The authors demonstrated that different microbial genera were recovered depending on the type of support material used, suggesting that the appropriateness of the support material in a microbial enrichment will depend on the microbial group of interest.
The DGGE analyses were performed as an efficient tool for the comparison of the microbial diversity recovered from the different bacterial enrichments implemented in this study. The results demonstrated that the polyurethane foams allowed the development of the most complex bacterial community, reflected by the higher number of dominant populations in the DGGE profiles, differently from the other enrichments, which yielded the lowest number of bands.
The composition of phytotypes at the phylum level was similar between the libraries derived from the WS (without support) and A (arenite) bacterial enrichments, both showing Thermotogae and Firmicutes as the predominant phyla. The phylum Thermotogae was the most predominant in the arenite library (96%), while in the WS clone library the Thermotogae and Firmicutes were found at the similar proportion (48.4% and 51.6% respectively). In many microbial community studies from oil reservoirs, injection water or other petroleum-associated environments, members of these two phyla are common inhabitants and, not rarely, predominant.2,41,42 Members of the family Thermotogaceae (order Thermotogales) belong to two physiological groups: extreme thermophiles that grow at temperatures above 70°C and moderate thermophiles that grow at lower temperatures.43 The genus Petrotoga, as the genera Geotoga and Thermotoga, is described as occurring exclusively in petroleum reservoirs.44,45
The S (shale) bacterial enrichment also yielded a community composed mainly by the phylum Thermotogae (94%), represented by the genus Petrotoga, followed by the phylum Proteobacteria (6%), represented only by the genus Rahnella. Although not commonly described in petroleum environments, this bacterium was previously reported as being involved in hydrocarbon biodegradation in Antarctic soils contaminated with polycyclic aromatic hydrocarbons (PAHs) and in enrichments from Brazilian petroleum samples.46,47 In a recent study of our research group,16 the bacterial diversity of the aerobic (AER) and anaerobic (ANA) enrichments of oil samples from Potiguar Basin (RN, Brazil) was evaluated by 16S rRNA clone library analysis, and 38.4% of the clones from the anaerobic enrichment were affiliated to the genus Rahnella.
The clone library originated from the PF (polyurethane foams) enrichments was more diverse at both genus and phylum levels. The use of polyurethane foam as support for the immobilization process has been already reported in literature, with wide application in studies of molecules such as enzymes,48 dyes49 and even building material.50 Polyurethane foam presents some features such as porosity, which does not only increase the surface area but also minimize the diffusion limitation for substrate and product.48 Silva et al.28 also described the use of polyurethane foams as an efficient support material for anaerobic biomass immobilization and increment of microbial growth, especially for sulfate reducing bacteria.
Phylotype analysis of the clone library derived from the PF enrichments revealed that the clones were affiliated to four different phyla in distinct abundances (Proteobacteria, Thermotogae, Firmicutes and Actinobacteria). Differences were more pronounced when comparing the microbiota recovered from the enrichments at the genus level. The PF enrichments allowed the recovery of representatives of 12 different genera, corroborating the efficiency of polyurethane foams for the improvement of the microbial diversity recovery. Many of the genera found using polyurethane foam as physical support, such as Geobacillus, Bacillus, Achromobacter, Acinetobacter, Pseudomonas, Kocuria and Paenibacillus, are described as organisms living in petroleum-associated environments, and some of them are involved in hydrocarbon degradation processes. The Geobacillus spp. constitute a thermophilic group, classified into the order Bacillales, described as being isolated from petroleum reservoirs.51 Liu and co-workers52 studied the alkB genes in species of this group, suggesting their potential as hydrocarbon degraders. The Achromobacter species have been previously described in the literature as hydrocarbon degraders and/or associated with oil field environments.22,53,54 A microbial diversity study conducted with injection water samples in platforms of the Campos Basin (Brazil) reported that 24% of the total 16S rRNA clones were related to the Achromobacter genus.55 The Pseudomonas, Bacillus and Acinetobacter spp. are commonly described as inhabitants of petroleum-associated environments, including reservoirs3,22,42,56–58 and their role is often linked to the hydrocarbon degradation process, including the production of biosurfactants.59,60 Similarly, the genera Kocuria and Paenibacillus have already been related to oil reservoirs61 and also to hydrocarbon degradation.62,63 The Paenibacillus spp. have been isolated from Iranian oil wells61 and the Kocuria from Chinese oil fields.64
The Leuconostoc is a bacterial group commonly described living in fresh plants and plays an important role in several industrial and food fermentation processes.65 Nonetheless, the presence of the genus Leuconostoc has already been reported in an oil field environment.21 Members of the genus Weissella are usually isolated from food and vegetables and have been involved with fermentative processes of food products.66 Recently, Silva and co-workers16 have found Kocuria, Bacillus, Weissella, Achromobacter, Acinetobacter and Leuconostoc in two different petroleum samples (Potiguar Basin, RN) from Brazilian reservoirs.
The genus Thermicanus was first described as a group encompassing thermophilic, fermentative microaerophilic bacteria living in soils.67 These bacteria were co-isolated together with a thermophilic acetogen, Moorella thermoacetica, from oxic soil obtained from Egypt, and these two species were shown to grow commensally on oligosaccharides via the interspecies transfer of H2 and formate and lactate.67 These data suggest that Thermicanus might be indirectly involved in the complex syntrophic degradation of hydrocarbons in oil-associated environments.
The results obtained using the different support materials, polyurethane foams, shale and arenite, revealed that all of them led to an increase of the microbial biomass, when compared to the enrichment without any physical support. However, it was expected that the use of shale and arenite would allow the recovery of a more diversified microbiota from oil samples, considering that they participate in the composition of the reservoir rock and are a natural support for biofilm formation in such environments. In fact, polyurethane foams yielded the highest microbial diversity. This could be explained by the porous nature of the polyurethane foams that allows an intense biofilm formation and EPS production, which in turn allows microorganisms that do not have affinity with the material surface to get attached to the first biofilm colonizers, thus increasing the microbial diversity.
Despite the diversity found in the anaerobic enrichments, members related to the sulfate reduction metabolism were not observed. This fact could be explained by the short period of incubation, 60 days, which could lead to an initial selection of facultative anaerobic heterotrophic bacteria, capable of degrading faster the nutrient source, becoming more abundant than sulfate-reducing members.
ConclusionThis work describes the use of three different physical supports – shale, arenite and polyurethane foams – for efficient biomass recovery in petroleum enrichments in sulfate reducing conditions. Molecular techniques and SEM were used as tools to assess the microbial diversity as a function of the physical support employed in the enrichments. Results revealed Petrotoga as the most abundant genus in the enrichments using shale and arenite as physical support. On the other hand, the enrichment using polyurethane foams was more diverse and allowed the identification of 12 different bacterial genera. Finally, the combined data gathered in this work demonstrated the usefulness of physical supports for the enrichment of low abundance microorganisms found in particular environments, such as deep oil reservoirs, enabling subsequent microbiological, physiological, genomic or metagenomic analyses.
Conflict of interestThe authors declare that there is no conflict of interest.
The authors thank Espaço da Escrita – Coordenadoria Geral da Universidade – UNICAMP – for the language services provided.