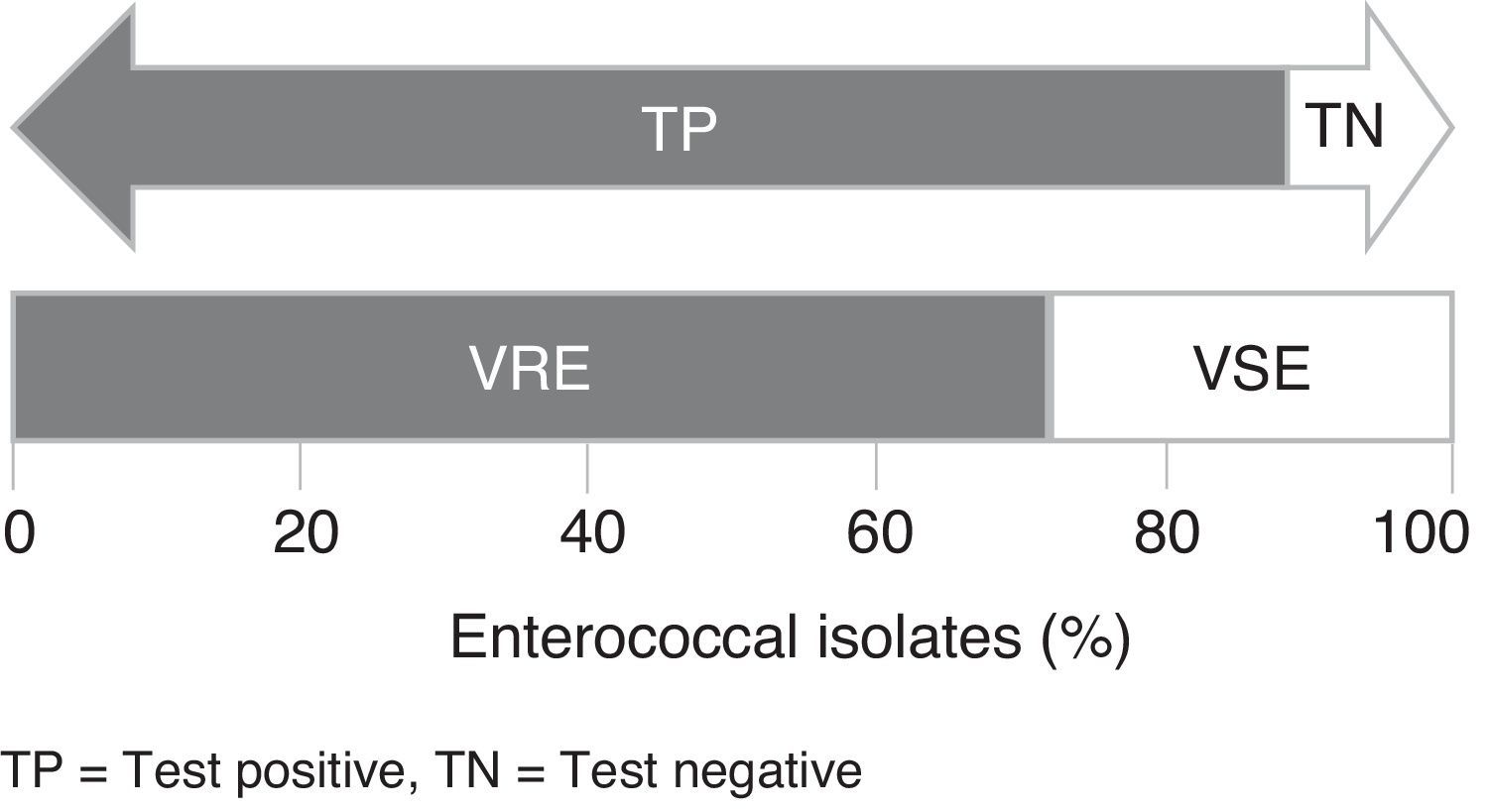
Rapid identification of vancomycin-resistant enterococci (VRE) can assist in choosing the appropriate treatment and preventing VRE spread. The performance of chromID™ VRE agar was evaluated using 184 clinical isolates of Enterococcus spp. and reference strains. The test had a sensitivity of 95.52% but a low specificity of 30%.
Vancomycin-resistant enterococci (VRE) are among the major agents of healthcare-associated infections and are considered a public health problem. Rapid VRE identification can assist in choosing the appropriate treatment and preventing VRE spread.1,2 The aim of this study was to evaluate the performance of a selective chromogenic medium for the detection and differentiation of vancomycin-susceptible and -resistant Enterococcus faecium and Enterococcus faecalis.
Vancomycin-susceptible enterococci (VSE) isolated from cases of infection (n=50) and VRE clinical isolates (n=134), including those collected from surveillance rectal swab cultures (n=62) and from cases of infection (n=72), were evaluated (Table 1). The following reference strains were also included: VRE strain (E. faecium, n=1) and VSE strains (E. hirae, n=1; E. gallinarum, n=2; E. faecium, n=1; and E. faecalis, n=4).
Clinical isolates used to evaluated the performance of the chromID™ VRE.
| Species | Susceptibility to vancomycina | Origin | ||
|---|---|---|---|---|
| S | R | Infection | Surveillance culture | |
| E. faecalis | 43 | 47 | 47 | 43 |
| E. faecium | 7 | 87 | 75 | 19 |
| Total | 50 | 134 | 122 | 62 |
All isolates were previously identified by phenotypic methods (hydrolysis of esculin in the presence of bile, production of pyrrolidonyl arylamidase, growth in broth containing 6.5% NaCl, and negative catalase test evidentiated by the absence of effervescence).3 Polymerase chain reaction was also used to confirm the presence of the genus Enterococcus and distinguish the species according to methods previously described by Ke et al.5 and Karyama et al.4 All isolates were obtained from the culture collection of the Gram-positive Cocci Laboratory – UFCSPA and stored in skim milk (Difco™) at −20°C. Vancomycin minimum inhibitory concentration was determined by broth microdilution according to CLSI guidelines (2015)6 and by Etest® according to the manufacturer's instructions. The chromID™ VRE (bioMérieux, Brazil S/A) assays were performed in two steps. First, the isolates that were previously stored in skim milk were grown in bile-esculin agar to confirm the presence of enterococci and check culture purity. Second, pure samples were grown in trypticase soy agar for 24h, followed by single-colony growth in chromID™ VRE agar at 37°C. After 24h, plates with any growth were considered VRE positive. A negative result was defined as a 48-h incubation period without any bacterial growth. According to the manufacturer's instructions, chromID™ VRE agar allows the identification of species based on the detection of enzyme activity. Therefore, E. faecium was stained purple and E. faecalis was stained blue-green.
Of the E. faecalis VRE tested (n=47), 41 were stained blue-green, 5 were stained gray, and 1 isolate did not grow. All E. faecium VRE tested (n=87) were stained purple. Among VSE isolates (n=50), 15 did not grow (10 E. faecalis and 5 E. faecium) and 35 (33 E. faecalis and 2 E. faecium) showed some growth at the edges of the plates in the corresponding color of each species, which may suggest false-positive results. All VSE-reference strains tested (n=8) did not show any visible growth in the chromogenic medium. Fig. 1 shows a schematic representation of the results obtained with chromID™ VRE agar.
The chromID™ VRE agar had a sensitivity of 87.23% and 100% for detecting E. faecalis VRE and E. faecium VRE, respectively, and a combined sensitivity and specificity of 95.52% and 30.00%, respectively, for detecting VRE. No difference was observed in the specificity and sensitivity at 24 and 48h. The positive predictive value (corresponding to the percentage of VRE that tested positive in chromID™ VRE) was 78.53% (95% confidence intervals; C.I.=72.22–84.83%) and the negative predictive value was 71.43% (95% C.I.=52.10–90.75%) (Table 2). Regarding sensitivity, similar results have been obtained in previous studies evaluating the performance of chromID™ VRE.1,7–14 In relation to the specificity, previous studies have obtained values higher than 95%,1,7,8,11 in contrast to the low specificity observed in this study.
Evaluation of chromID™ VRE in detecting true positive vancomycin-resistant enterococci.
| ChromID™ VRE agar | Gold standard (VRE)a | Gold standard (VSE)a | Total |
|---|---|---|---|
| Test positive | 128 | 35 | 163 |
| Test negative | 6 | 15 | 21 |
| Total | 134 | 50 | 184 |
| Sensitivity | 95.52% (95% C.I.=92.02–99.02%) | ||
| Specificity | 30.00% (95% C.I.=17.29–42.70%) | ||
| Positive predictive value | 78.53% (95% C.I.=72.22–84.83%) | ||
| Negative predictive value | 71.43% (95% C.I.=52.10–90.75%) | ||
| Accuracy | 77.72% (95% C.I.=71.70–83.73%) | ||
Colonies with non-discriminatory staining (gray, dark, or colorless) have been reported in some studies.10,15 We also observed VRE colonies with a grayish shade, which may lead to false-negative results.
Among all VRE isolates, 100% of the E. faecium grew within 24h. The same was observed in previous studies.11,14 According to Grabsch et al.,8 24-h identification of VRE allows earlier confirmation of colonization by these strains, facilitates infection control, and helps to avoid the spread of microorganisms. However, one E. faecalis VRE isolate did not grow even after 48h, which may suggest that some strains can exhibit a different behavior and/or require more time to grow in the medium.
Delmas et al.1 compared growth before and after an enrichment step in bile-esculin agar supplemented with vancomycin in order to select only VRE strains. The enrichment step improved the performance of chromID™ VRE at 24h of incubation. Other studies have shown that strains incubated overnight in an enrichment broth containing vancomycin as a first step followed by the use of chromID™ VRE resulted in improved specificity or sensitivity.1,10,11,14,16 However, this method is only useful for fecal specimens due to the large number of different microorganisms that can be present in these samples.
Most studies evaluating chromID™ VRE performance have used only fecal specimens (stool samples and rectal swabs) or only resistant strains. In our study, we evaluated well-characterized vancomycin-resistant and -susceptible isolates in order to observe the possible occurrence of false-positive results, because incorrectly prescribed antibiotics have a negative clinical impact. Despite the data presented here, a possible limitation of this study is that fecal samples were not included, because the density of microorganisms in a clinical specimen may affect the correct diagnosis.
In conclusion, the chromID™ VRE agar is a rapid and useful tool for the screening and identification of VRE, with a good sensitivity of about 96.00%. However, because specificity (30.00%) was limited by false positive VRE mainly, we recommend further VRE identification by conventional tests to avoid misinterpretation.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and Universidade Federal de Ciências da Saúde de Porto Alegre. We also thank the bioMérieux, Brazil S/A for the supply of the chromID™ VRE agar plates.