The main objective of the present study was to isolate and characterize actinomycetes for their plant growth-promotion in chickpea. A total of 89 actinomycetes were screened for their antagonism against fungal pathogens of chickpea by dual culture and metabolite production assays. Four most promising actinomycetes were evaluated for their physiological and plant growth-promotion properties under in vitro and in vivo conditions. All the isolates exhibited good growth at temperatures from 20°C to 40°C, pH range of 7–11 and NaCl concentrations up to 8%. These were also found highly tolerant to Bavistin, slightly tolerant to Thiram and Captan (except VAI-7 and VAI-40) but susceptible to Benlate and Ridomil at field application levels and were found to produce siderophore, cellulase, lipase, protease, chitinase (except VAI-40), hydrocyanic acid (except VAI-7 and VAI-40), indole acetic acid and β-1,3-glucanase. When the four actinomycetes were evaluated for their plant growth-promotion properties under field conditions on chickpea, all exhibited increase in nodule number, shoot weight and yield. The actinomycetes treated plots enhanced total N, available P and organic C over the un-inoculated control. The scanning electron microscope studies exhibited extensive colonization by actinomycetes on the root surface of chickpea. The expression profiles for indole acetic acid, siderophore and β-1,3-glucanase genes exhibited up-regulation for all three traits and in all four isolates. The actinomycetes were identified as Streptomyces but different species in the 16S rDNA analysis. It was concluded that the selected actinomycetes have good plant growth-promotion and biocontrol potentials on chickpea.
Chickpea (Cicer arietinum L.) is the second most widely grown food legume crop after common bean, with annual production of 13.8mt worldwide.1 Globally, more than 90% of chickpea production occurs in the semi-arid tropics of Asia and Africa. Asia accounts for 88% of global chickpea production whereas India is the largest producer accounting for 75% of Asia's chickpea production.2 Several biotic and abiotic factors were involved in low production of chickpea. The biotic factors include fungi, bacteria, viruses, nematodes, mycoplasma and insect pests. Fungi are the largest group affecting stems, roots, leaves, flowers and pods. Chickpea crop is mainly affected by Fusarium wilt, dry root rot, collar rot, Ascochyta blight and Botrytis gray mold caused by Fusarium oxysporum f. sp. ciceri (FOC), Macrophomina phaseolina, Sclerotium rolfsii, Ascochyta rabiei and Botrytis cinerea, respectively resulting in reduced crop yield.3,4
The microbes in rhizosphere help plants in growth-promotion and yield. Actinomycetes are one of the major components of rhizosphere microbial populations and are useful in soil nutrient cycling5,6 as well as plant growth-promotion (PGP).7 Actinomycetes produce secondary metabolites such as lytic enzymes, PGP substances and antibiotics.8 The actinomycetes, mainly those belonging to Streptomyces spp., make up an important group of soil microbes. Streptomyces are abundant in soil and help in the degradation of complex molecules to simple molecules for plant growth and development.9,10 These are also reported to decompose organic matter, promote plant growth and control plant pathogens.11
In the present study, actinomycetes isolated from rhizosphere and herbal vermicompost were characterized and evaluated for PGP properties and for biocontrol-related traits. The promising strains were also evaluated for their PGP in chickpea under field conditions.
Materials and methodsActinomycetes isolationActinomycetes were isolated from herbal vermicompost (Jatropha curcas, Annona squamosa, Parthenium hysterophorus, Gliricidia sepium and Azadirachta indica) and chickpea rhizosphere soils. Ten grams of each vermicompost and rhizosphere soils were suspended in 90mL of sterile physiological saline (0.85% NaCl in distilled water) in a bottle and kept for shaking on an orbital shaker (at 100rpm) at 28±2°C for 1h. At the end of shaking, the samples were serially diluted up to 105 dilutions and samples from 104 and 105 dilutions were spread plated (0.1mL) on actinomycetes isolation (AIA) agar (HiMedia Laboratories, Mumbai, India) and incubated at 28±2°C for 7 days. Prominent colonies were isolated and stored on AI agar slants.
Selection of antagonistic actinomycetes against fungal pathogens of chickpeaA total of 89 actinomycetes were screened for their antagonistic activity against S. rolfsii, M. phaseolina (three strains viz. MP-6, MP-24 and MP-115) and FOC (acquired from legumes pathology, ICRISAT, Patancheru) by dual culture assay as per the protocols of Gopalakrishnan et al.12 on glucose casaminoacid yeast extract agar plates. The culture filtrates of the promising isolates, based on the dual culture assay, were extracted by partitioning against ethyl acetate (EtOAc) and the resultant organic (EtOAc) and aqueous fractions were evaporated on a rotary evaporator and collected in a minimal volume of methanol. Both the fractions were evaluated for their antagonistic potential against the three fungal pathogens of chickpea (M. phaseolina, FOC and S. rolfsii). For this, a fungal disc of 6mm diameter was bored and kept at center of the potato dextrose agar plate amended with either organic or aqueous fractions (at a concentration of 0.5%). Control plates contained only methanol. The plates were incubated at 28±2°C for 5 days and inhibition of the pathogen was recorded for both dual culture and metabolite production assays on a scale of 0–4 as follows: 0=no inhibition; 1=slight inhibition; 2=moderate inhibition; 3=good inhibition and 4=excellent inhibition.
Colony morphology of selected actinomycetesSelected isolates of actinomycetes were streaked on AI agar by quadrant streaking technique and incubated for 5 days at 28±2°C. At the end of incubation, the isolated colonies were observed for their morphology. Gram staining was also performed13 and observed under light microscope.
Molecular identification of the selected actinomycetesThe selected actinomycetes were sent to Macrogen Inc., Seoul, Korea for identification by 16S rDNA analysis. The sequences obtained from Macrogen were compared with similar sequences from GenBank, compared using the BLAST program,14 aligned using the Clustal W software15 and the dendrogram inferred by neighbor-joining method.16 Bootstrap analysis was performed using the MEGA version 4 program to estimate the statistical stability of the branches in cluster with 1000 replications. The sequences (1460bp for SAI-13, 1474bp for SAI-291,475bp for VAI-7 and 1472bp for VAI-40) were submitted to NCBI and accession numbers obtained.
In vitro evaluation of actinomycetes for their physiological traits and fungicide tolerancePhysiological properties such as tolerance to pH, temperature, salinity and fungicides were studied for the selected actinomycetes. The actinomycetes were streaked on Bennett's agar (HiMedia Laboratories, Mumbai, India), adjusted to different pH (5, 7, 9 and 11) and saline concentrations (0–12% NaCl at the interval of 2%) and incubated at 28±2°C for 5 days. For temperature, the Bennett's agar plates were streaked with the actinomycetes and incubated at different temperatures (20°C, 30°C, and 40°C) for 5 days. For test at 50°C, the isolates were inoculated in Bennett's broth and incubated at 50°C. The fungicide tolerance was evaluated as per the protocols of Gopalakrishnan et al.17 The actinomycetes were streaked on AI agar plates amended with fungicides Bavistin, Thiram, Benlate, Captan and Ridomil at field application levels (2500, 3000, 4000, 3000 and 3000ppm) and incubated at 28±2°C for 5 days. At the end of 5 days incubation the growth of the actinomycetes were rated. The rating scale for pH, temperature and salinity were recorded on a scale of 0–3 as follows: 0=no growth; 1=slight growth; 2=moderate growth and 3=good growth.
Evaluation for production of extracellular enzymes, siderophore, indole acetic acid (IAA) and hydrocyanic acid (HCN)Selected actinomycetes were evaluated for their PGP and biocontrol traits including siderophore, chitinase, cellulase, lipase, protease, HCN, β-1,3-glucanase and IAA production. Siderophore production was estimated as per the protocol of Schwyn and Neilands.18 Chitinase production was estimated by amending agar plates with colloidal chitin suspension and mineral salts according to the standard protocols of Hsu and Lockwood.19 Production of cellulase was detected by using standard protocols of Hendricks et al.20 The protease and lipase productions were estimated as per the protocols of Bhattacharya et al.21 HCN was qualitatively assessed by the method described by Lorck.22 Estimation of IAA was done as per the protocols of Patten and Glick.23 The rating scales for siderophore, chitinase, cellulase, lipase and protease were as follows: 0=no halo zone; 1=halo zone of 1–10mm; 2=halo zone of 11–20mm; 3=halo zone of 21–30mm; 4=halo zone of 31–40mm; and 5=41–50mm. For HCN production, the following rating scale was used: 0=no color change, 1=light reddish brown, 2=medium reddish brown and 3=dark reddish brown.
Evaluation of PGP potential of actinomycetes by “ragdoll” methodGrowth-promoting activity was evaluated by the ‘ragdoll’ method as described by Chamblee and Green.24 In brief, chickpea seeds (ICCV 2) were surface sterilized by using 2.5% sodium hypochlorite solution for 5min and washed thoroughly with sterilized distilled water. Sterilized seeds were soaked in individual actinomycetes (108colony forming units (CFU)mL−1) for 1h and placed in one half of a wet paper towel, folded and rolled into a moderately tight tube and placed in a plastic bag. This tube was kept at (28±2°C) for 5 days. At the end of incubation, % germination, root and shoot lengths were measured.
Evaluation of PGP potential of actinomycetes under field conditionsThe selected actinomycetes were evaluated for PGP potential on chickpea under on-station field conditions in 2013–14 post-rainy season at ICRISAT Patancheru (17°30′N; 78°16′E; altitude=549m), Hyderabad, Telangana, India. Chickpea variety ICCV 2 was used for this trial. Soils at the experimental site were of vertisol which contained 25% sand, 21% silt and 52% clay with alkaline pH of 7.5–8.5. The organic C content of this field was 0.56%. The rhizosphere soil (top 15cm) contained 642ppm of total N and 9.03ppm of available P. The experiment was laid out in a randomized complete block design with three replicates with a plot size of 4m×3 ridges. The actinomycete cultures were grown in SCB at 28±2°C for 5 days. Chickpea seeds were treated with the actinomycete cultures (108CFUmL−1) for 50min and sown on 2nd November 2013 at a row-to row spacing of 60cm and a plant-to-plant spacing of 10cm. The actinomycete cultures (1000mL; 108CFUmL−1) were also applied once in 15 days to the soil close to the plant until flowering stage. The control plots contained no actinomycete strains. No serious phytopathogens or insect pest attacks were observed during the cropping period. Irrigation was done on 23 days after sowing (DAS) and 51 DAS whereas weeding on 22 DAS and 49 DAS. At 60 DAS, the nodule number, stem weight, pod number, pod weight, leaf weight and leaf area were noted and compared with un-inoculated control. The crop was harvested manually on 4th Feb 2014 and at harvest, stover yield, grain yield and total dry matter were noted. Rhizosphere soil samples were collected from a depth of 0–15cm at both flowering (60 DAS) and crop maturity stages and analyzed for total N (ppm), available P (ppm) and organic C % according to the protocols of Novozamsky et al.,25 Olsen and Sommers,26 and Nelson and Sommers,27 respectively and soil biological properties including microbial biomass C, microbial biomass N and dehydrogenase activities as per the protocols of Anderson and Domsch,28 Brooks et al.,29 and Casida,30 respectively.
Colonization studiesChickpea roots were examined for colonization by actinomycetes under scanning electron microscope (SEM) at RUSKA lab, College of Veterinary Science, Rajendranagar, Hyderabad, Telangana, India, as per the protocols of Bozzola and Russell.31 For this, the seeds of chickpea (ICCV 2) were surface sterilized as explained earlier and allowed to sproutovernight. The sprouted seeds were soaked in selected actinomycetes for 50min and transferred carefully into sand tubes containing sterilized coarse sand (50g). Booster dose of actinomycete (1mL; 108CFUmL−1) was applied after 7 days. The tubes were incubated at 24±2°C in a light chamber with an average illumination of 9600lx and photosynthetic photon flux of 350μEm−2s−1. After two weeks of incubation, chickpea seedlings were taken out and the roots were washed in 0.1M phosphate buffer. Root tips of 4–5mm length were cut and fixed in glutaraldehyde (2.5%) in phosphate buffer for 24h at 4°C. At the end of 24h incubation, the root samples were again washed with phosphate buffer, post fixed in osmium tetraoxide (2%) for 4h and dehydrated using a graded series of ethanol. The dehydrated samples were dried, with a critical-point liquid carbon dioxide as a transition fluid, and adhered onto aluminum specimen mounts with double stick adhesive tape. The mounted samples were coated with gold-palladium in an automated sputter coater (JEOL JFC-1600) and examined under SEM (JOEL-JSM 5600).
Gene expression studiesThe selected actinomycetes were grown in Bennett's broth for 72h. RNA was extracted from actinomycetes by conventional Trizol method.32 The quality and quantity of RNA was estimated by Nanodrop (Thermo Scientific, USA) and RNA integrity by 2100 Bioanalyzer (Agilent, USA). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed as per the manufacturer's instructions using Applied Biosystems 7500 Real Time PCR System with the SYBR green chemistry (Applied Biosystems, USA). Gene specific primers for IAA, siderophore and β-1,3-glucanase were designed using Primer3 software.33 Primers for IAA (F: GTCACCGGGATCTTCTTCAAC; R: GATGTCGGTGTTCTTGTCCAG); siderophore (F: ATCCTCAACACCCTGGTCTG; R: TCCTTGTACTGGTACGGGACTT) and β-1,3-glucanase (F: CCGAACACCACCTACTCCAC; R: CCAGGTTGAGGATCAGGAAG) were designed from the sequences collected from UniProtKB database (http://www.uniprot.org/uniprot) as described in Gopalakrishnan et al.34 RNA polymerase principal sigma factor HrdB (SCO5820) (F: GGTCGAGGTCATCAACAAGC; R: CTCGATGAGGTCACCGAACT) was used as the endogenous control. qRT-PCR reactions and conditions were conducted as described earlier.34 PCR reactions were carried out in 10μL reactions containing 30ng of first strand cDNA, 1X PCR buffer, 125mM dNTPs, 1.5mM MgCl2, 0.2mM primers and 1U Taq polymerase and PCR cycle was programmed as, 50°C for 2min and denaturation at 95°C for 10min followed by 40 cycles of denaturation at 95°C for 15s and annealing and extension at 60°C for 1min. The data obtained from different PCR runs or cDNA samples was analyzed using the mean of the CT values of the three biological replicates that were normalized to the mean CT values of the endogenous gene. The expression ratios were calculated using the 2−ΔΔCt method and relative transcription levels were plotted graphically.
Statistical analysisData were analyzed by using analysis of variance (ANOVA) technique, by SAS GLM (General Linear Model) procedure (SAS Institute 2002-08, SAS version 9.3) considering isolates and replication as fixed in randomized complete block design. Isolate means were tested for significance and compared using Fisher's protected least significant difference.
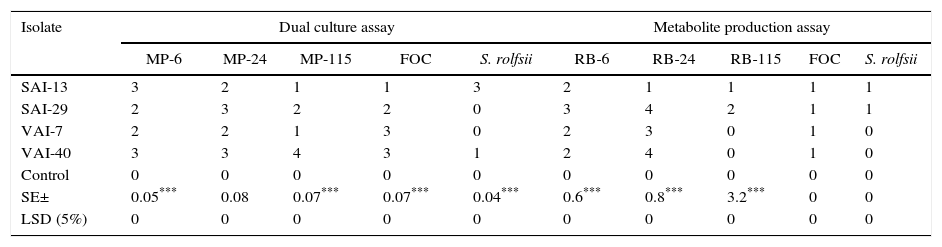
ResultsScreening for potential actinomycetesOf the 89 isolates screened for their antagonistic potential against important pathogens of chickpea such as FOC, M. phaseolina and S. rolfsii by dual culture assay, four actinomycetes viz. SAI-13 and SAI-29 (isolated from chickpea rhizosphere), VAI-7 (from A. squamosa vermicompost) and VAI-40 (from J. curcas vermicompost) were found to be most promising. When the culture filtrates of these four isolates were partitioned against EtOAc, by solvent extraction method, only organic fractions were found active. Among the four isolates, the organic fractions of SAI-13 and SAI-29 were found to be potential against all the fungal pathogens (Table 1).
Antagonistic potential of the four actinomycetes against fungal pathogens of chickpea.
| Isolate | Dual culture assay | Metabolite production assay | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MP-6 | MP-24 | MP-115 | FOC | S. rolfsii | RB-6 | RB-24 | RB-115 | FOC | S. rolfsii | |
| SAI-13 | 3 | 2 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 |
| SAI-29 | 2 | 3 | 2 | 2 | 0 | 3 | 4 | 2 | 1 | 1 |
| VAI-7 | 2 | 2 | 1 | 3 | 0 | 2 | 3 | 0 | 1 | 0 |
| VAI-40 | 3 | 3 | 4 | 3 | 1 | 2 | 4 | 0 | 1 | 0 |
| Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SE± | 0.05*** | 0.08 | 0.07*** | 0.07*** | 0.04*** | 0.6*** | 0.8*** | 3.2*** | 0 | 0 |
| LSD (5%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
MP-6, MP-24 and MP-115 – three strains of Macrophomina phaseolina; FOC, Fusarium oxysporum f. sp. ciceri; S. rolfsii, Sclerotium rolfsii. The rating scale 0=no inhibition, 1=slight inhibition, 2=moderate inhibition, 3=good inhibition, 4=excellent inhibition. LSD, least significant difference; SE, standard error.
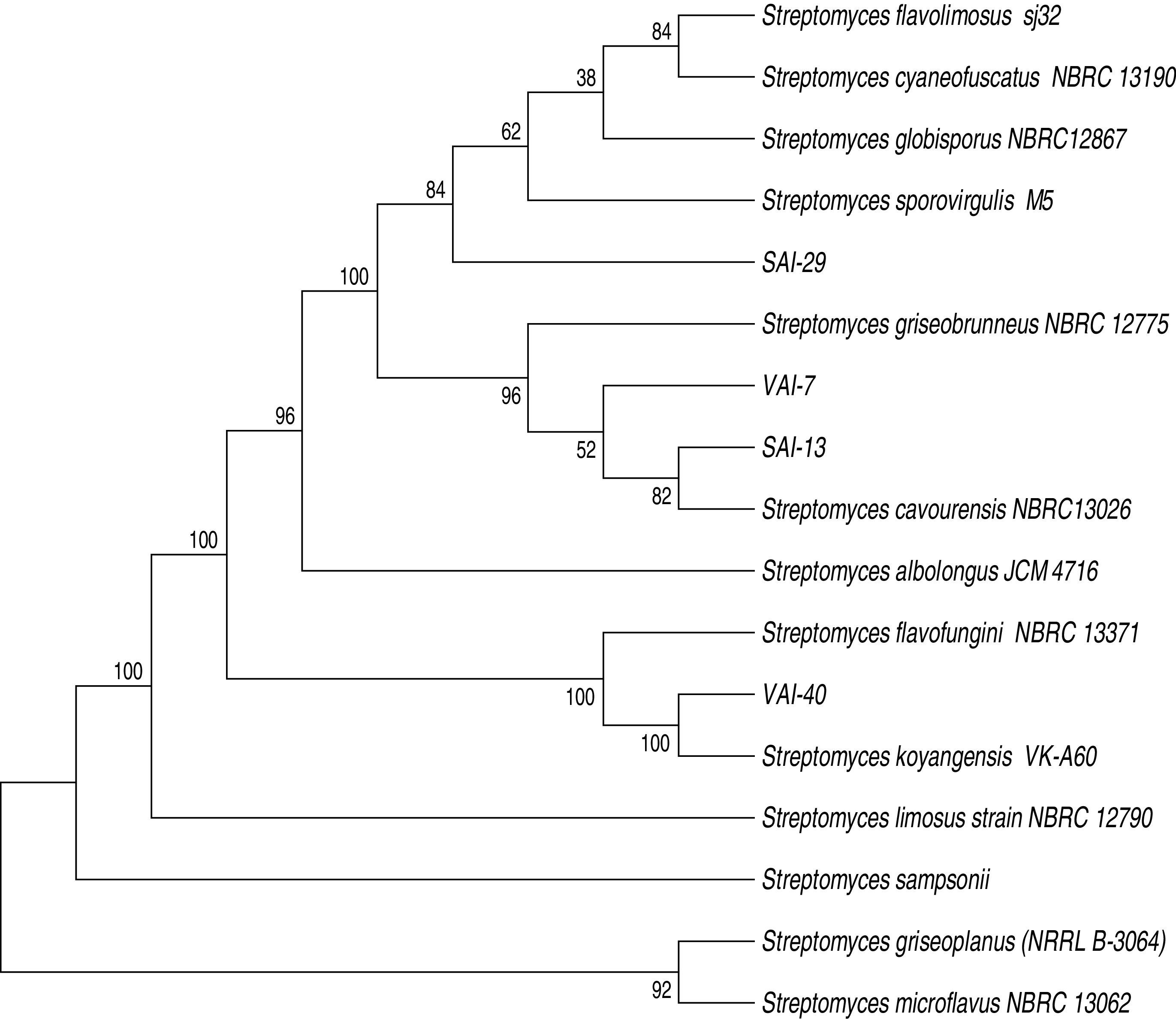
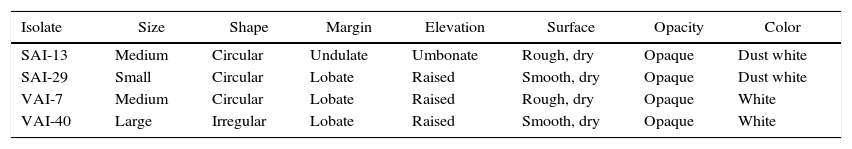
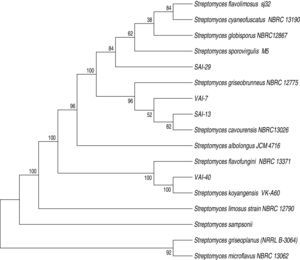
All the four isolates were found to be Gram positive and the colonies showed sporulating aerial mycelium. The colony morphology description of the four isolates is presented (Table 2). Analysis of 16S rDNA sequences by neighbor-joining method revealed that all four isolates matched to Streptomyces spp. (100%). The nucleotide sequences were submitted to GenBank and NCBI accession numbers were obtained as follows: SAI-13: KM220609, SAI-29: KM220608, VAI-7: KM220610; and VAI-40: KM220611 (Fig. 1).
Individual colony morphology of four actinomycetes on AIA.
| Isolate | Size | Shape | Margin | Elevation | Surface | Opacity | Color |
|---|---|---|---|---|---|---|---|
| SAI-13 | Medium | Circular | Undulate | Umbonate | Rough, dry | Opaque | Dust white |
| SAI-29 | Small | Circular | Lobate | Raised | Smooth, dry | Opaque | Dust white |
| VAI-7 | Medium | Circular | Lobate | Raised | Rough, dry | Opaque | White |
| VAI-40 | Large | Irregular | Lobate | Raised | Smooth, dry | Opaque | White |
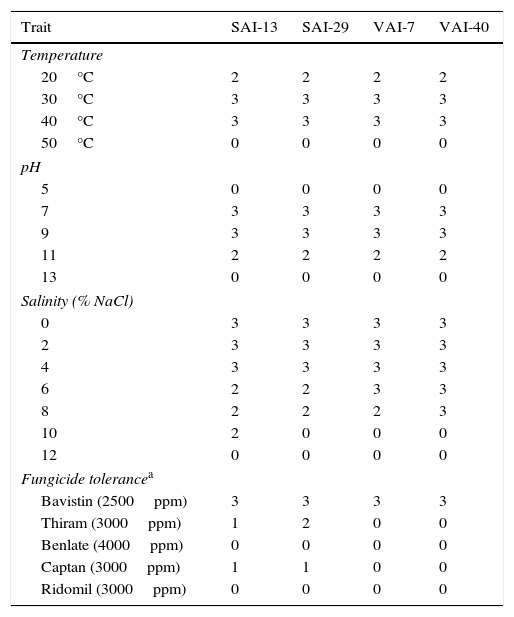
All the four actinomycetes grew between pH 7 and 11, but maximum sporulation was observed at pH 9 while no growth was observed at pH 5. The actinomycete isolates also grew well at temperatures between 20°C and 40°C but unable to grow at 50°C and all isolates tolerated NaCl concentration of 8% (SAI-13 tolerated up to 10%). When the isolates were tested for fungicide tolerance, all the four isolates were found tolerant to Bavistin and susceptible to Benlate and Ridomil whereas two of them, SAI-13 and SAI-29, were found moderately tolerant to Thiram and slightly tolerant to Captan (Table 3).
Effect of temperature, pH, salinity and fungicides on the growth of four actinomycetes.
| Trait | SAI-13 | SAI-29 | VAI-7 | VAI-40 |
|---|---|---|---|---|
| Temperature | ||||
| 20°C | 2 | 2 | 2 | 2 |
| 30°C | 3 | 3 | 3 | 3 |
| 40°C | 3 | 3 | 3 | 3 |
| 50°C | 0 | 0 | 0 | 0 |
| pH | ||||
| 5 | 0 | 0 | 0 | 0 |
| 7 | 3 | 3 | 3 | 3 |
| 9 | 3 | 3 | 3 | 3 |
| 11 | 2 | 2 | 2 | 2 |
| 13 | 0 | 0 | 0 | 0 |
| Salinity (% NaCl) | ||||
| 0 | 3 | 3 | 3 | 3 |
| 2 | 3 | 3 | 3 | 3 |
| 4 | 3 | 3 | 3 | 3 |
| 6 | 2 | 2 | 3 | 3 |
| 8 | 2 | 2 | 2 | 3 |
| 10 | 2 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 |
| Fungicide tolerancea | ||||
| Bavistin (2500ppm) | 3 | 3 | 3 | 3 |
| Thiram (3000ppm) | 1 | 2 | 0 | 0 |
| Benlate (4000ppm) | 0 | 0 | 0 | 0 |
| Captan (3000ppm) | 1 | 1 | 0 | 0 |
| Ridomil (3000ppm) | 0 | 0 | 0 | 0 |
0=no growth; 1=slight growth; 2=moderate growth; 3=good growth.
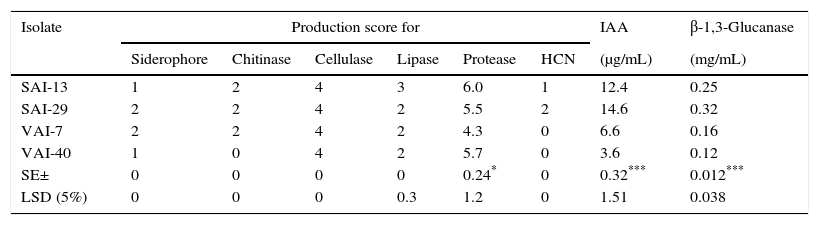
Under in vitro conditions, all the four actinomycetes were found to produce siderophore, cellulase, lipase, protease, IAA, β-1,3-glucanase, chitinase (except VAI-40) and HCN (except VAI-40 and VAI-7). Of the four isolates, SAI-29 produced higher amounts of HCN, IAA, and β-1,3-glucanasewhile SAI-13 produced higher amounts of lipase and protease (Table 4).
Production of extracellular enzymes and plant growth promoting traits by four actinomycetes.
| Isolate | Production score for | IAA | β-1,3-Glucanase | |||||
|---|---|---|---|---|---|---|---|---|
| Siderophore | Chitinase | Cellulase | Lipase | Protease | HCN | (μg/mL) | (mg/mL) | |
| SAI-13 | 1 | 2 | 4 | 3 | 6.0 | 1 | 12.4 | 0.25 |
| SAI-29 | 2 | 2 | 4 | 2 | 5.5 | 2 | 14.6 | 0.32 |
| VAI-7 | 2 | 2 | 4 | 2 | 4.3 | 0 | 6.6 | 0.16 |
| VAI-40 | 1 | 0 | 4 | 2 | 5.7 | 0 | 3.6 | 0.12 |
| SE± | 0 | 0 | 0 | 0 | 0.24* | 0 | 0.32*** | 0.012*** |
| LSD (5%) | 0 | 0 | 0 | 0.3 | 1.2 | 0 | 1.51 | 0.038 |
HCN, hydrocyanic acid; IAA, indole acetic acid. The rating scales for siderophore, chitinase, cellulase, lipase and protease were as follows: 0=no halo zone, 1=halo zone of 1–10mm, 2=halo zone of 11–20mm, 3=halo zone of 21–30mm, 4=halo zone of 31–40mm; 5=41–50mm. For HCN production, the following rating scale was used: 0=no color change, 1=light reddish brown, 2=medium reddish brown, 3=dark reddish brown. LSD, least significant difference; SE, standard error.
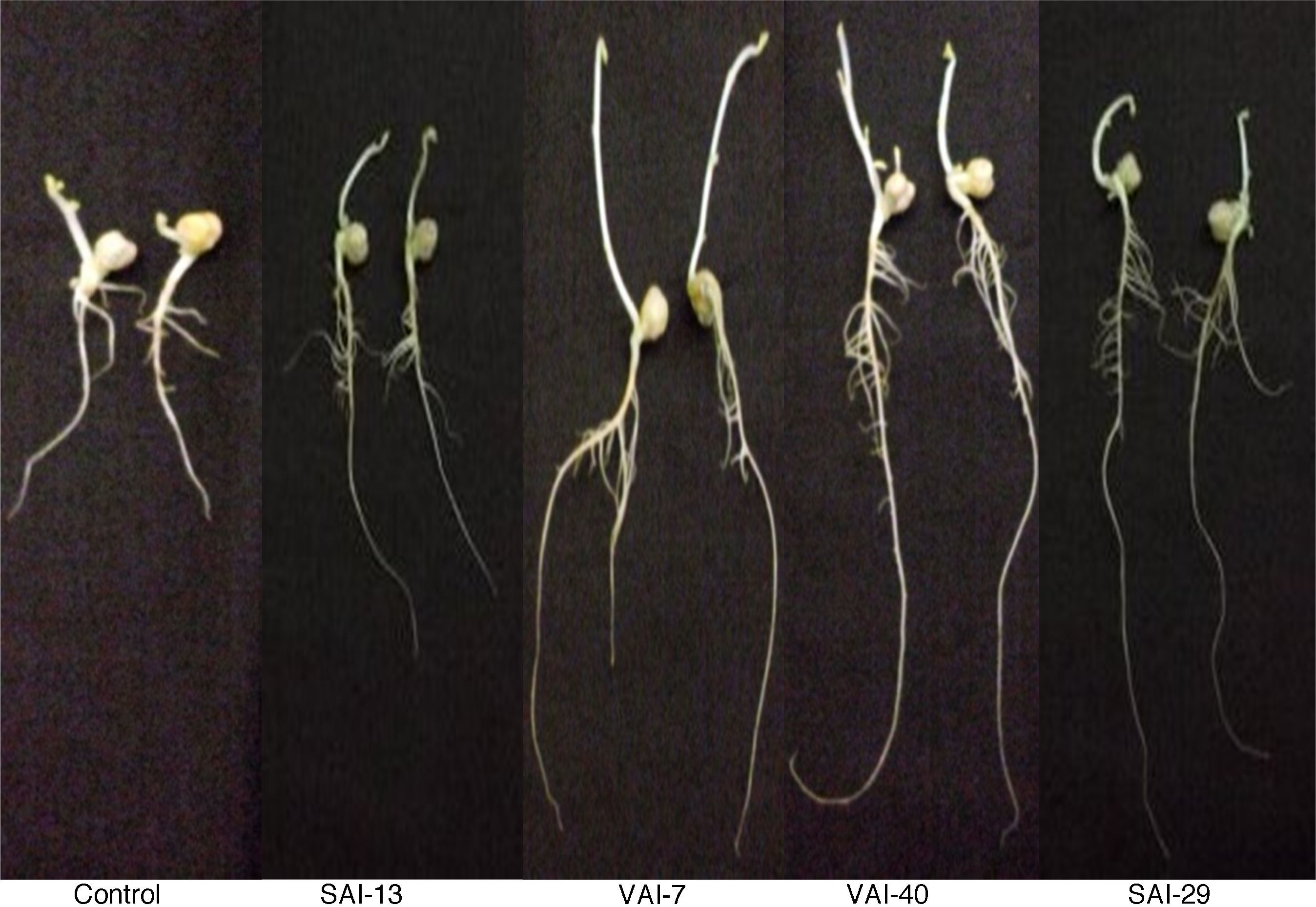
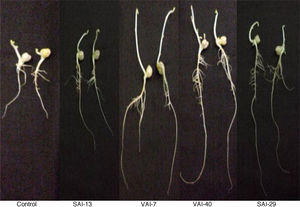
The effect of actinomycetes on chickpea growth was demonstrated by “ragdoll” method in which all isolates exhibited increase in root (17%) and shoot (3%) lengths when compared with the control. Of the four actinomycetes tested, VAI-7 significantly enhanced both root and shoot lengths whereas VAI-40 and SAI-29 significantly enhanced only root lengths when compared to control (Fig. 2).
Under field conditions, a considerable increase in agronomic and soil mineral properties were observed in actinomycetes treated plots over control plots. At 60 DAS, there was an increase in nodule number (up to 19%), leaf area (up to 27%), pod number (up to 26%), leaf weight (up to 16%), plant height (up to 3%) and stem weight (up to 16%). While at final harvest, the actinomycetes treated plots exhibited enhanced the stover yield (up to 23%), grain yield (up to 20%), total dry matter (up to 17%), seed weight (up to 16%) and seed number (up to 22%) (Table 5). The rhizosphere soil from actinomycetes treated plots enhanced up to 11% of total N, 27% of available P, 8% of organic C, 33% of dehydrogenase activity and 22% of microbial biomass carbon, at flowering stage and 3% of total N, 19% of available P, 11% of organic C, 26% of dehydrogenase activity and 29% of microbial biomass carbon at harvest when compared with the control (Table 6).
Effect of the four actinomycetes on agronomic performance and yield potential of chickpea under field conditions – at crop maturity.
| Isolate | At 60 days after sowing | At crop harvest | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nodule number(plant−1) | Leaf area(cm−2plant−1) | Leaf weight(gplant−1) | Pod number(plant−1) | Shoot weight(gplant−1) | Plant height(cm) | Seed weight(gplant−1) | Seed number(plant−1) | Stover yield(tha−1) | Grain yield(tha−1) | Total dry matter(tha−1) | |
| SAI-13 | 50 | 660 | 4.13 | 62 | 4.17 | 48 | 196 | 77 | 1.70 | 1.70 | 3.40 |
| SAI-29 | 59 | 665 | 3.82 | 64 | 3.99 | 49 | 199 | 80 | 2.17 | 1.85 | 4.01 |
| VAI-7 | 60 | 692 | 4.41 | 79 | 4.69 | 49 | 202 | 70 | 1.89 | 2.09 | 3.98 |
| VAI-40 | 61 | 855 | 4.09 | 61 | 4.05 | 48 | 196 | 85 | 1.72 | 1.82 | 3.53 |
| Control | 49 | 626 | 3.71 | 59 | 3.96 | 47 | 195 | 66 | 1.68 | 1.67 | 3.34 |
| SE± | 2.1** | 36.4** | 0.134* | 3.2** | 0.159* | 0.4* | 0.448** | 1.6*** | 0.059*** | 0.089* | 0.129** |
| LSD (5%) | 6.7 | 118.8 | 0.436 | 10.4 | 0.490 | 0.6 | 3.7 | 5.2 | 0.175 | 0.249 | 0.346 |
LSD, least significant difference; SE, standard error.
Effect of the four actinomycetes on rhizosphere soil mineral properties of chickpea under field conditions (2013–2014) –at flowering and crop maturity.
| Isolate | At flowering | At crop maturity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total N(ppm) | Available P(ppm) | Organic carbon(%) | Dehydrogenase activity(μgTPFg−1 soil24h−1) | Microbial carbon(μgg−1 soil) | Total N(ppm) | Available P(ppm) | Organic carbon(%) | Dehydrogenase activity(μgTPFg−1 soil24h−1) | Microbial carbon(μgg−1 soil) | |
| SAI-13 | 759 | 9.6 | 0.56 | 61.5 | 634 | 725 | 7.1 | 0.56 | 76.0 | 584 |
| SAI-29 | 685 | 9.3 | 0.56 | 43.5 | 649 | 729 | 7.1 | 0.56 | 71.0 | 778 |
| VAI-7 | 742 | 12.6 | 0.59 | 50.5 | 777 | 736 | 8.5 | 0.59 | 74.5 | 608 |
| VAI-40 | 726 | 9.5 | 0.57 | 60.0 | 782 | 747 | 7.5 | 0.62 | 68.0 | 568 |
| Control | 675 | 9.2 | 0.54 | 41.0 | 612 | 721 | 6.9 | 0.55 | 55.0 | 551 |
| SE± | 11.9* | 0.26** | 0.005** | 2.37** | 29.7* | 4.6* | 0.14** | 0.01* | 2.51* | 24.9** |
| LSD (5%) | 46.8 | 1.03 | 0.018 | 9.31 | 116.7 | 15.3 | 0.52 | 0.038 | 9.87 | 97.9 |
LSD, least significant difference; SE, standard error; TPF: triphenylformazan.
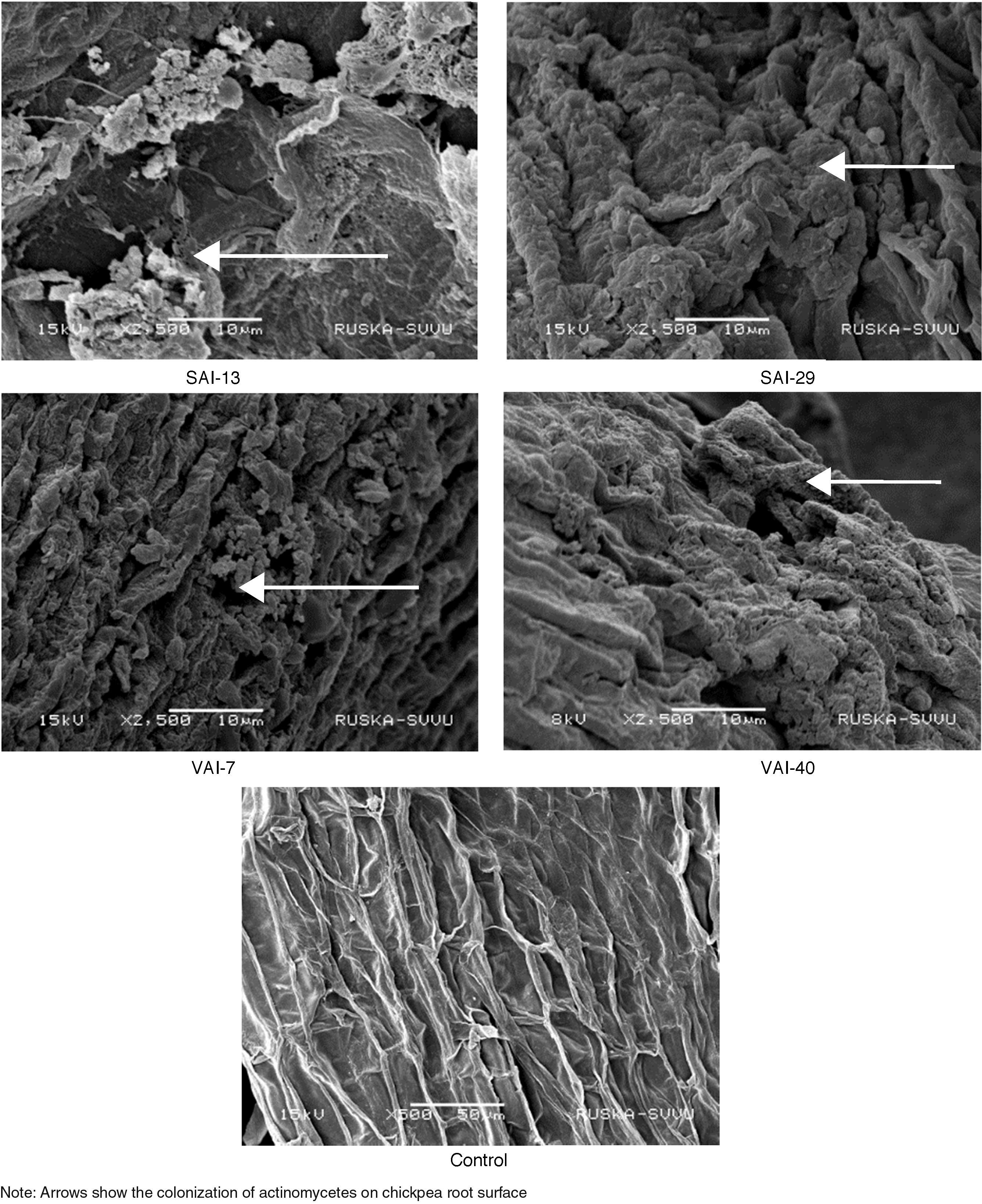
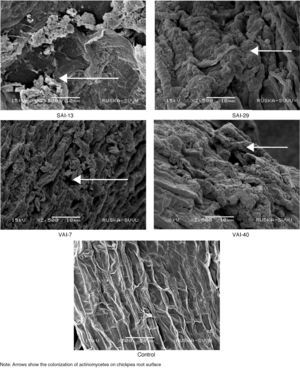
Colonization of actinomycetes on chickpea roots was confirmed by images of SEM in actinomycetes treated roots. All the isolates exhibited good colonization on chickpea roots without any damage to root surface. The hyphae of Streptomyces were also seen to grow and adhere to the surface of the root (Fig. 3).
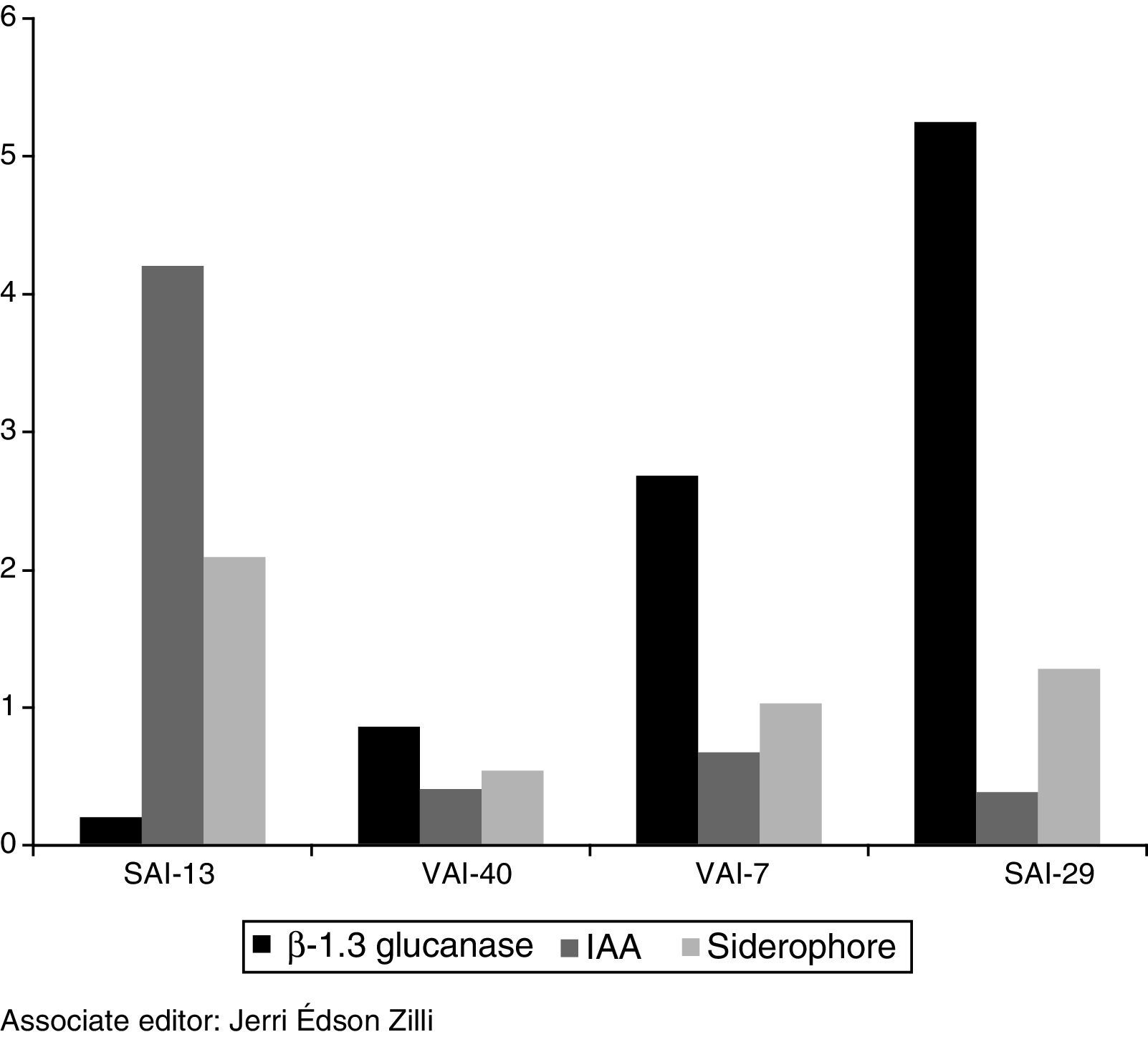
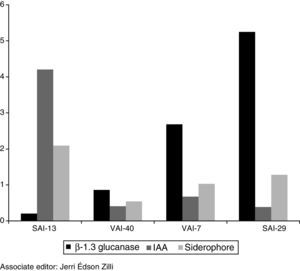
In PGP gene expression profile studies, good quality RNA was isolated from all the four actinomycete strains. qRT-PCR analysis on selected PGP genes of actinomycetes revealed up-regulation of all the three genes and on all the four actinomycetes. β-1,3-Glucanase gene was highly up-regulated in SAI-29 (5 folds) followed by VAI-7, VAI-40 and SAI-13 while IAA gene was highly up-regulated in SAI-13 (4 folds) followed by VAI-7, VAI-40 and SAI-29. Siderophore gene was highly up-regulated in SAI-13 (1 fold) followed by SAI-29, VAI-7 and VAI-40 (Fig. 4).
DiscussionActinomycetes are used for PGP and biocontrol of plant pathogens in agriculture. For instance, Streptomyces spp. were reported to enhance PGP of crops such as tea,35 wheat,36 rice,34 bean,11 tomato37 and pea.38 In the present study, when the 89 actinomycetes screened for their antagonistic potentials against important fungal pathogens of chickpea. Of which, four isolates (SAI-13, SAI-29, VAI-7 and VAI-40) were found to be promising. There was no inhibition of S. rolfsii by SAI-29 in dual culture assay however, when the pathogen reached the antagonist the growth of the pathogen was arrested. Hence, SAI-29 was further selected for metabolite production assay in which the organic fraction was found to be effective in controlling S. rolfsii. Similarly, VAI-40 was found to be effective in dual culture assay while not effective in metabolite production assay, perhaps the metabolite produced was of volatile in nature. Of the four isolates, SAI-13 and SAI-29 were found to have broad spectrum inhibition against all tested pathogens of chickpea. These four isolates found to vary in their shape, elevation, opacity, margins, color and size. Molecular identification of the four isolates, based on their 16S rDNA sequence analysis, revealed that all actinomycetes belong to Streptomyces spp. Similar observations were noted by Araújo39 where he observed Streptomyces as dry, smooth or hairy colonies with airborne mycelium of different colors. However, these phenotypic characteristics of Streptomyces vary with the composition of the culture medium.40
In the present investigation, all the Streptomyces strains grew well at temperatures 20°C to 40°C, an alkaline pH of 11 and saline concentrations of up to 8%. Hence, it can be concluded that the Streptomyces isolates used in this study can be used in semi-arid tropics, alkaline and saline soils. The distribution of Streptomyces spp. in diverse ecosystems is one of the main reasons for ability to adapt to a wide range of environmental conditions, allowing them to survive in high temperatures, saline, acidic and alkaline soils.39 These traits help the Streptomyces spp. in competing with the native microflora when introduced into the rhizosphere of the host.41 The salinity and pH levels of the rhizosphere soil are important factors for the microbes to compete and survive in rhizosphere. Therefore, the tolerance for high salinity and pH should be the criteria for selection of microorganisms.42
In this study, all the four Streptomyces spp. were tolerant to Bavistin whereas SAI-13 and SAI-29 were tolerant to Thiram and Captan at field application levels. These fungicides were generally used to control soilborne fungal pathogens. Hence, it is concluded that the four Streptomyces spp. are compatible with Bavistin, Thiram and Captan and hence can be treated together on the seeds to control soil borne fungal pathogens and thus reducing the use of fungicides in controlling pathogens.
The selected four Streptomyces spp. possess biocontrol as well as PGP properties such as production of IAA, siderophore, β-1,3-glucanase, lipase, protease, cellulase, chitinase (except VAI-40) and HCN (except VAI-7 and VAI-40). IAA is a phytohormone which comes under auxins group and helps in improving plant growth by stimulating cell elongation, root initiation, seed germination and seedling growth.43 Siderophores are usually produced by various soil microbes including Streptomyces spp. to bind Fe3+ from the environment and make it available for its own growth; plants also utilize these as an iron source.44Streptomyces spp. from rhizosphere soil has been reported to produce siderophores and inhibit the growth of phytopathogens.38 HCN production is also reported to play a role in disease suppression.45 Some of the recent studies indicated that metabolites produced by microbes such as HCN may enhance plant establishment in rhizosphere.46 The isolates from the rhizosphere soil of chickpea were observed to exhibit HCN production potential which promoted plant growth directly or indirectly or synergistically.47 Suwan et al.48 proposed that actinomycetes act as potential as biological control agents because of its intense antagonistic activity via the production of various antifungal substances such as chitinase and β-1,3-glucanase.
In the present investigation, the effect of the four Streptomyces spp. on the growth of chickpea seedlings was studied by “ragdoll” method, in which VAI-7 exhibited maximum increase in shoot and root lengths. This observation was further confirmed in the field trials. Under field conditions, the Streptomyces spp. increased nodule number over un-inoculated control demonstrating a direct proof for enhancing nitrogen fixation. The Streptomyces strains used in this study exhibited increase in agronomic properties such as the shoot weight, leaf weight, leaf area, plant height, grain yield and stover yield over the un-inoculated control. They also increased the total N, available P, organic C %, microbial biomass C, microbial biomass N and dehydrogenase properties in plots treated with Streptomyces spp. The phosphate mineralization is mainly provided by phosphatases or phosphohydrolases. These enzymes convert the unavailable phosphate to available form by slow mineralization of phytic acid and poly phosphates.49 In the present study all the actinomycetes produced lytic enzymes indicating their ability to convert complex molecules to simpler available forms, which may be attributed to the increased levels of total N and available P in the inoculated plots. The substrate used in measuring the dehydrogenase activity is 2,3,5 triphenyltetrazolium chloride (TTC) and was converted to triphenylformazan (TPF) by the action of dehydrogenase, which is directly proportional to the number of microorganisms in soil.28 In the present study, all the plots exhibited increase in TPF concentration when compared with the un-inoculated control plots indicates that the Streptomyces spp. applied in the field were able to survive in the rhizosphere and confer PGP. Among the four strains of Streptomyces used in this study, VAI-7 was found to enhance highest grain yield and total dry matter (by 20% and 17%, respectively). The enhancement in yield by VAI-7 could be its both direct and indirect PGP traits including IAA, siderophore, chitinase, cellulase, lipase, β-1,3-glucanase and protease (Table 4). Further, as mentioned earlier, VAI-7 also enhanced highest shoot as well as root lengths of chickpea seedlings (Fig. 2). VAI-7 was also found to enhance greatly soil biological and mineral nutrient properties including microbial biomass carbon, dehydrogenase, total N, available P and organic carbon (Table 6). Hence, the Streptomyces strain VAI-7 could be exploited for its PGP and yield enhancement traits.
The SEM images, in this study, showed that all the four Streptomyces strains colonized the roots of chickpea without causing any damage. Compounds such as aromatic and phenolic compounds, sugars, amino acids, organic acids and carbohydrates from root exudates acts as chemo attractants that helps in root colonization.50,51 Colonization accounts to the proliferation of microbes but not their movement.1 Weller52 reported that a microorganism that colonizes root is ideal for use as biocontrol as well as plant growth-promoting agents.53 Actinomycetes strains like Micromonospora spp., Streptomyces spp., Streptosporangium spp., and Thermobifida spp., were reported as best to colonize the plant rhizosphere, showing an immense potentiality as biocontrol agent against wide range of root pathogenic fungi.54 Association of actinomycetes confers many advantages to plants like production of antibiotics, extracellular enzymes, phytohormones, siderophores and phosphate solubilization, protects plant against biotic and abiotic stress.55 It is concluded that all the tested Streptomyces strains not only colonized on the roots but also proliferated and enhanced PGP in chickpea. In the present study, gene expression studies for IAA, siderophore and β-1,3-glucanase genes exhibited up-regulation of the three genes, supporting the biochemical results. The importance of gene expression has been widely recognized in gene function.56
Conflicts of interestThe authors declare no conflicts of interest.
We thank Council of Scientific and Industrial Research, New Delhi, India, for the financial support provided to M Sreevidya during her PhD. This work was undertaken as part of the CGIAR Research Program on Grain Legumes. ICRISAT is a member of CGIAR Consortium. We would also like to thank ICRISAT and all of the staff members of the biocontrol unit, including Alekhya G, Satya A, Prasad PVS, Manohar P, Nagappa B, Bharath D and Jabbar A for their significant inputs in the laboratory and field experiments.