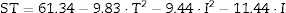Biosurfactants have many advantages over synthetic surfactants but have higher production costs. Identifying microorganisms with high production capacities for these molecules and optimizing their growth conditions can reduce cost. The present work aimed to isolate and identify a fungus with high biosurfactant production capacity, optimize its growth conditions in a low cost culture medium, and characterize the chemical structure of the biosurfactant molecule. The fungal strain UFSM-BAS-01 was isolated from soil contaminated with hydrocarbons and identified as Fusarium fujikuroi. To optimize biosurfactant production, a Plackett–Burman design and a central composite rotational design were used. The variables evaluated were pH, incubation period, temperature, agitation and amount of inoculum in a liquid medium containing glucose. The partial structure of the biosurfactant molecule was identified by nuclear magnetic resonance spectrometry. F. fujikuroi reduced surface tension from 72 to 20mNm−1 under the optimized conditions of pH 5.0, 37°C and 7 days of incubation with 190rpm agitation. The partial identification of the structure of the biosurfactant demonstrated the presence of an α,β-trehalose. The present study is the first report of the biosynthesis of this compound by F. fujikuroi, suggesting that the biosurfactant produced belongs to the class of trehalolipids.
Biosurfactants are secondary metabolites produced by microorganisms under specific growth conditions. Biosurfactants have an amphiphilic structure, enabling them to reduce surface and interfacial tension in water–oil and oil–water systems. Biosurfactants can be classified into several classes: glycolipids, lipopeptides, lipoproteins, phospholipids, fatty acids, polymeric biosurfactants and particulate biosurfactants.1 Their structural diversity allows biosurfactants to perform a variety of functions in the petrochemical, environmental, pharmaceutical, food, and agricultural industries, among others.2
Compared to synthetic surfactants, biosurfactants present lower toxicity, higher biodegradability, greater resistance to extreme environmental conditions, are produced from renewable sources and have greater ecological acceptability.3 Despite these advantages, a limitation to the industrial use of biosurfactants is their higher production costs compared to synthetic surfactants.4 One means of reducing these costs is to identify and use microorganisms with high production capacities for these molecules. Within the diversity of known microorganisms, few are good producers of biosurfactants. Some fungi can produce larger amounts of biosurfactants than bacteria, which is explained by their cell wall stiffness.5
Another way to reduce the cost of biosurfactants is to optimize growth medium conditions for the producing microorganisms. Several environmental factors influence biosurfactant yield and quality, particularly the carbon and nitrogen source, pH, aeration, inoculum quantity and incubation period.6–8 The present work aimed to isolate and identify a fungus with high biosurfactant production capacity from soil samples contaminated with hydrocarbons, to optimize its growth conditions in a low cost culture medium and to characterize the chemical structure of the biosurfactant molecule by nuclear magnetic resonance spectrometry techniques.
Material and methodsFungus isolationMicroorganisms were isolated from 10 soil samples contaminated with hydrocarbons from mechanical workshops and fuel stations in the city of Santa Maria, RS, Brazil (29° 41′ 03″S, 53° 48′ 25″ W). One gram of soil was added to a mineral medium containing type B diesel [with 6% (v/v) biodiesel] as the sole source of carbon and energy. The mineral medium had the following macronutrient composition (gL−1): 0.04 CaCl2·2H2O; 0.1 KH2PO4; 0.8 NaCl; 1.0 NH4Cl; 0.2 MgSO4·7H2O; 0.1 KCl; and micronutrients (mgL−1): 0.1 CoCl2·6H2O; 0.425 MnCl2·4H2O; 0.05 ZnCl2; 0.015 CuSO4·5H2O; 0:01 NiCl2·6H2O. The pH was adjusted to 5.8. The diesel oil (10mLL−1) was filtered through a 0.22μm membrane and mixed with medium that had been previously autoclaved at 121°C for 20min. The culture medium containing the diesel oil and the soil was incubated at 30°C and 120rpm for 7 days. An aliquot of 1mL was transferred every seven days to the same sterile medium and incubated under the same conditions. After seven transfers, 1mL of the resulting medium was diluted to 10−6 and a 0.1mL aliquot of each dilution was added to Petri dishes containing PDA medium at pH 5.8. The plates were incubated for 96h at 30°C in a microbiological oven.
Selection of biosurfactant-producing fungusFrom the 10 soil samples, five isolates were obtained with different colony morphologies. The fungi were purified from sequential replicates using PDA medium. To select the best biosurfactant producers, fungi were grown in five 250mL Erlenmeyer flasks containing 50mL of liquid culture medium with the following composition (gL−1): 30.0 glucose; 1.0 NH4NO3; 6.0 KH2PO4; 2.7 Na2HPO4;0.1 MgSO4·7H2O; 0.0012 CaCl2; 0.00165 FeSO4·7H2O; 0.0015 MnSO4·4H2O and 0.0022 Na-EDTA. The vials were incubated for 6 days at 120rpm and 32°C. Every two days, the culture medium samples were centrifuged at 10,000rpm for 4min, and the supernatant was collected for the measurement of the surface tension (mNm−1) and emulsification index. The surface tension was assessed by the pendant drop method on 10 drops of supernatant using a DSA 25E goniometer (KrüssGmbH, Hamburg, Germany). The emulsification index was evaluated by mixing 2mL of the supernatant with 2mL of filtered diesel oil in flat bottom test tubes and vortexing for 40s. The emulsification index (% IE24) was determined as described by Nitschke and Pastore9 by the division between the height of the emulsion layer and the total height of the solution, as measured by digital electronic calliper.
Fungus identificationThe most promising fungus for the production of biosurfactants was coded as UFSM-BAS-01 and identified by partial sequencing of nuclear ribosomal DNA (nrDNA). The fungus was cultivated in potato-dextrose (PD) liquid medium, and its genomic DNA was extracted using a ZR MiniPrep® ZR fungi/bacteria kit (Zymo Research, Irvine, CA, USA). Elongation factor 1α (EF-1α) is often used to investigate the genus Fusarium.10 An amplification reaction for the target fragment (∼700bp) was performed following the methods of O’Donnell et al.11 PCR products were purified using a GenElute PCR cleaning kit® (Sigma, St. Louis, USA) following the manufacturer's instructions. Sequencing of the samples was performed on the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The sequenced fragments were analysed using the 2.0.0b Staden package program12 to obtain the consensus sequence for EF-1α and BLASTn was performed. The sequence was deposited in GenBank.
The phylogenetic relationships of the samples were carried out by EF region sequences aligned in the program BioEdit version 7.2.513 and reconstructed based on MEGA 5.0 software14 with Maximum Likelihood (ML) analysis for a total of 1000 replicates for all reconstructions. The Tamura-Nei nucleotide substitution model with Gamma distribution was estimated using FindModel software.15 The sequences most related to that obtained in the present work were selected to construct a cladogram of the genus from the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/), including Neonectria radicicola as the outgroup.
Optimization of biosurfactant productionA Plackett–Burman (PB) design was used to optimize biosurfactant production by the fungus in liquid medium with glucose as the only source of carbon and energy. The variables evaluated were pH (5.0–7.0), incubation time (2–7 days), temperature (28–37°C), agitation (50–190rpm) and amount of inoculum (1–3 disks of fungal mycelium). Table 1 shows the levels investigated for each variable in the PB design, comprising 12 experiments and three central points. Based on the interpretation of the results, a central composite rotational design (CCRD) was used for the significant PB variables: incubation time (2, 5, 7, 9 and 12 days), agitation (50, 90, 120, 150 and 190rpm) and temperature (37, 39, 42, 45 and 47°C).
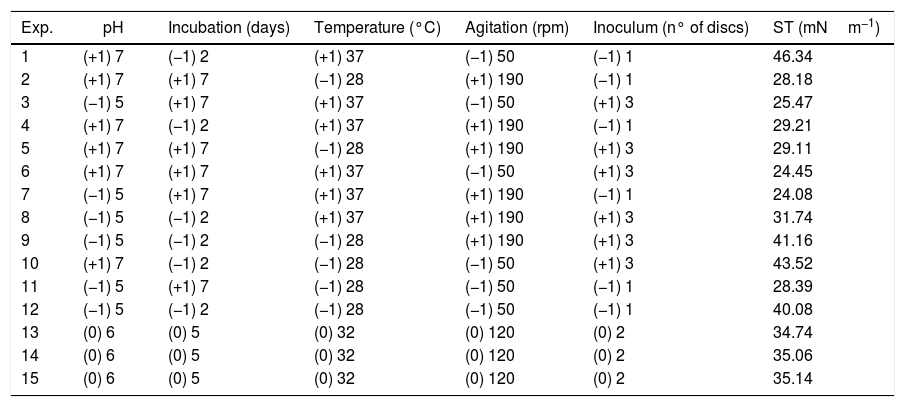
Surface tension (ST) values of the cell-free culture medium (supernatant) after the growth of the fungus Fusarium fujikuroi UFSM-BAS-01 under different environmental conditions, combined through a Plackett–Burman (PB) design.
| Exp. | pH | Incubation (days) | Temperature (°C) | Agitation (rpm) | Inoculum (n° of discs) | ST (mNm−1) |
|---|---|---|---|---|---|---|
| 1 | (+1) 7 | (−1) 2 | (+1) 37 | (−1) 50 | (−1) 1 | 46.34 |
| 2 | (+1) 7 | (+1) 7 | (−1) 28 | (+1) 190 | (−1) 1 | 28.18 |
| 3 | (−1) 5 | (+1) 7 | (+1) 37 | (−1) 50 | (+1) 3 | 25.47 |
| 4 | (+1) 7 | (−1) 2 | (+1) 37 | (+1) 190 | (−1) 1 | 29.21 |
| 5 | (+1) 7 | (+1) 7 | (−1) 28 | (+1) 190 | (+1) 3 | 29.11 |
| 6 | (+1) 7 | (+1) 7 | (+1) 37 | (−1) 50 | (+1) 3 | 24.45 |
| 7 | (−1) 5 | (+1) 7 | (+1) 37 | (+1) 190 | (−1) 1 | 24.08 |
| 8 | (−1) 5 | (−1) 2 | (+1) 37 | (+1) 190 | (+1) 3 | 31.74 |
| 9 | (−1) 5 | (−1) 2 | (−1) 28 | (+1) 190 | (+1) 3 | 41.16 |
| 10 | (+1) 7 | (−1) 2 | (−1) 28 | (−1) 50 | (+1) 3 | 43.52 |
| 11 | (−1) 5 | (+1) 7 | (−1) 28 | (−1) 50 | (−1) 1 | 28.39 |
| 12 | (−1) 5 | (−1) 2 | (−1) 28 | (−1) 50 | (−1) 1 | 40.08 |
| 13 | (0) 6 | (0) 5 | (0) 32 | (0) 120 | (0) 2 | 34.74 |
| 14 | (0) 6 | (0) 5 | (0) 32 | (0) 120 | (0) 2 | 35.06 |
| 15 | (0) 6 | (0) 5 | (0) 32 | (0) 120 | (0) 2 | 35.14 |
The fungus was cultured under the optimized conditions for 7 days. Cells were removed by centrifugation at 10,000rpm for 4min, and the supernatant was membrane filtered with a pore size of 0.22μm. The cell-free supernatant was acidified to pH 4.0 using 6M HCl and held overnight for precipitation.2 Then, 50mL of the supernatant, 50mL of hexane (to remove fatty acids) and 50mL of chloroform were added followed by an equal volume of ethyl acetate:methanol (1:4) at room temperature. A compound in the form of transparent crystals (C1) was isolated upon extraction with ethyl acetate:methanol.
The remaining extract was basified with 4M ammonium hydroxide and subjected to a new extraction procedure with the solvents chloroform, ethyl acetate and methanol (2:4). A powdery brown substance was obtained upon ethyl acetate and methanol extraction, which was subjected to a new extraction process with chloroform, ethyl acetate and n-butanol. In the fraction with n-butanol, a white crystalline (C2) compound with characteristics of sugars was isolated and subjected to nuclear magnetic resonance spectrometry, as described below.
NMR spectrometry and melting point1H and 13C NMR spectra were recorded on a 600MHz nuclear magnetic resonance spectrometer (Bruker, Magneto Ascend 600 Console Avance III HD, Germany). The (uncorrected) melting point values of the substances were determined on a digital melting point apparatus (Microchemistry, model MQAPF-301, Brazil).
GC-FID/GC–MS analysisThe samples were analysed by GC-FID and GC–MS. The autosampler used was an AOC-20is series injector (Shimadzu, Japan), the gas chromatograph coupled to the flame ionization detector (GC-FID) was a GC-2010 Plus (Shimadzu, Japan), and the gas chromatograph coupled to the mass spectrometer detector was a GCMS-QP2010 Ultra (Shimadzu, Japan). The composition was elucidated by comparison with an analytical standard of methylated fatty acid ester – FAME mix standard (Supelco, Bellefonte, PA, USA). Individual components were identified using their relative retention indices with the Wiley Registry of Mass Spectral Data (Palisade Corporation, Newfield, NY, USA).
Determination of the critical micellar concentration (CMC)The cell-free supernatant was kept overnight at room temperature. After the addition of ammonium sulphate (40%, w/v), the supernatant was centrifuged at 10,000rpm for 4min. The precipitate was extracted twice with ice-cold acetone, and the crude biosurfactant was collected as dry powder after the evaporation of the acetone. The CMC was determined by adding concentrations of 2.5–150mgL−1 of the crude biosurfactant in distilled water.2
ResultsIsolation, selection and identification of the fungusFrom the 10 soil samples, five biosurfactant-producing isolates were obtained. In the first test, UFSM-BAS-01 was distinguished from other fungi by its higher biosurfactant production, its reduction of the surface tension of the culture medium from 72 to 52mNm−1 and its higher emulsification index (24.4%).
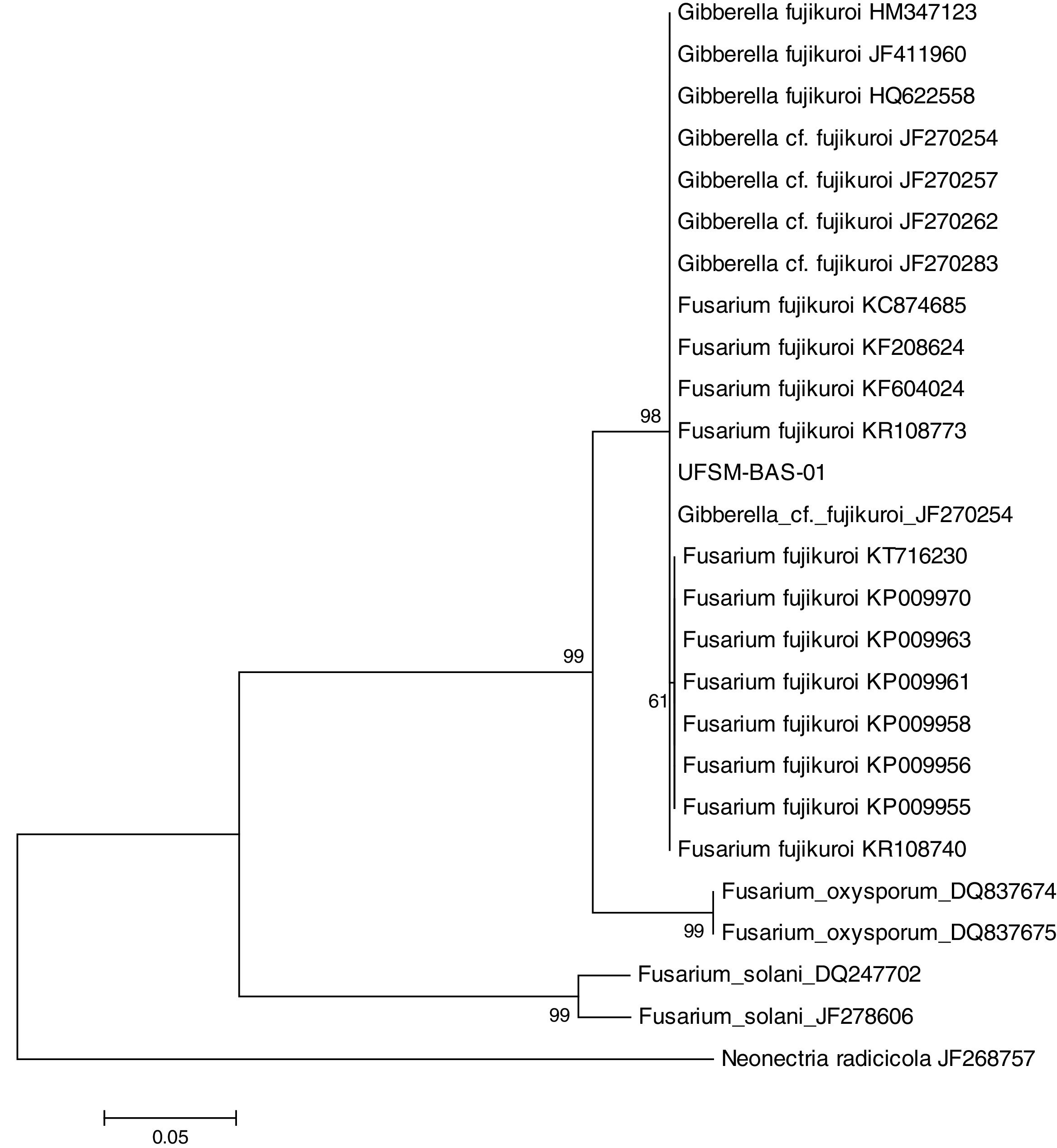
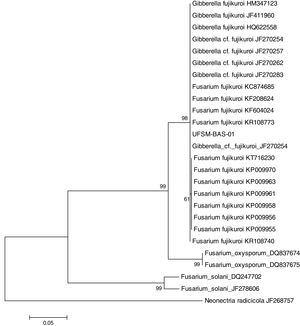
This isolate was identified with the help of molecular tools. Using a comparative analysis by BLASTn in NCBI, the consensus sequence showed 100% similarity with two species: Fusarium fujikuroi16 and Gibberella fujikuroi17 (Fig. 1). However, these are two names for the same fungus because F. fujikuroi is the anamorph phase and G. fugikuroi is the teleomorph phase of the same organism.18 Three subtypes were verified by ML analysis. F. fujikuroi and G. fujikuroi were grouped in the F. fujikuroi species complex,19 and the other two clades identified were Fusarium oxysporum and Fusarium solani. Therefore, the elongation factor 1α (EF-1α) was highly informative and was able to identify the isolate as belonging to the G. fujikuroi species complex, with 99% bootstrap support. The consensus sequence was deposited in GenBank under accession number: KX574231.
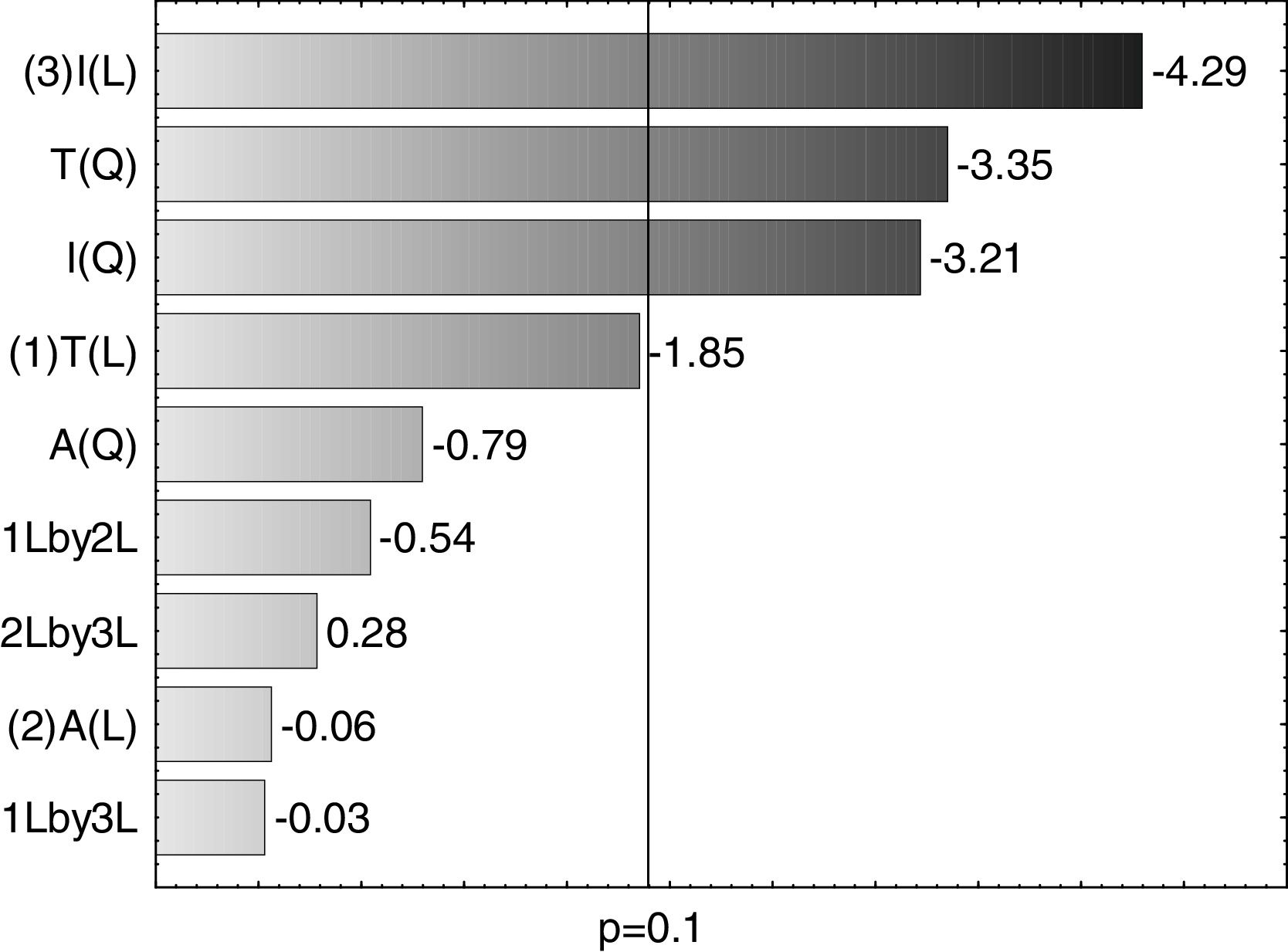
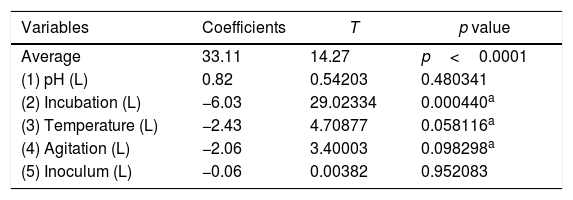
Optimization of biosurfactant productionThe optimization of the culture conditions for the production of a biosurfactant in mineral medium plus glucose was performed using a PB matrix (Table 1). The results were validated by analysis of variance (ANOVA) and the coefficient of determination (R2) was 0.8071. The lowest surface tension value was obtained in experiment 7 (24.08mNm−1). The temperature, agitation and incubation time variables significantly affected the surface tension (Table 2).
Linear regression coefficients for the reduction of surface tension after the growth of the fungus Fusarium fujikuroi UFSM-BAS-01 under different environmental conditions, combined through a Plackett–Burman (PB) design.
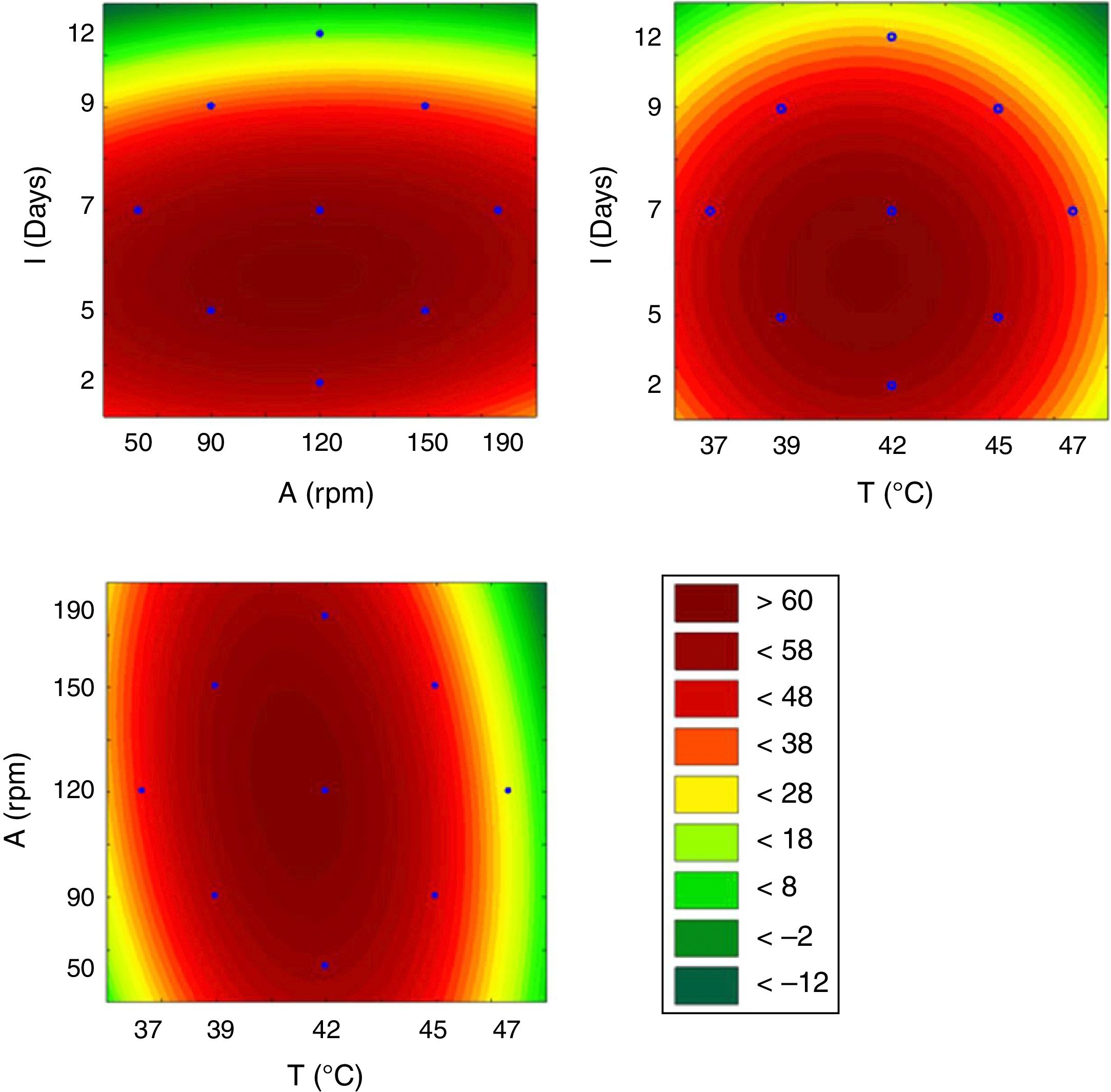
From the results obtained with the PB design, a central rotational compound (CCRD) was designed with 17 experiments to optimize the statistically significant variables from the PB matrix: incubation, temperature and agitation. In PB, the minimum value of ST was 24.08mNm−1, while in CCRD the minimum measured value was 20.08mNm−1, in conditions of 47°C, 120rpm for 7 days of incubation. The highest production of the biosurfactant by the fungus F. fujikuroi UFSM-BAS-01 occurs at thermophilic temperatures, even though the fungus was isolated under mesophilic conditions.
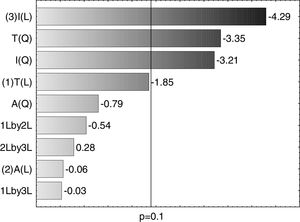
A Pareto graph (Fig. 2) represents the significant variables in the CCRD with a significance level of 90% (p<0.1). Confirming previous observations with the PB matrix, lower ST occurred at higher temperatures and longer incubation periods. However, unlike PB, the agitation variable was not statistically significant, which also justified the performance of the CCRD.
The following equation describes the behaviour of surface tension (ST) in response to temperature (T) and incubation time (I):
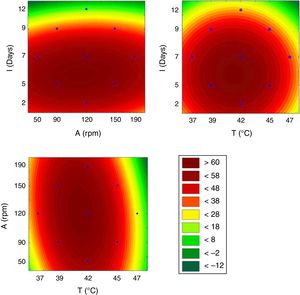
This model was validated by analysis of variance (ANOVA), and the coefficient of determination (R2) was 0.84953. A graphical representation of the optimization of biosurfactant production demonstrates that the lowest values of ST (dark green) were obtained at the positive axial point for incubation and at the central point for agitation (Fig. 3a). As shown in Fig. 3b, longer incubation periods and higher temperatures provided lower STs. As shown in Fig. 3c, the higher the temperature and the agitation, the lower the ST of the culture medium.
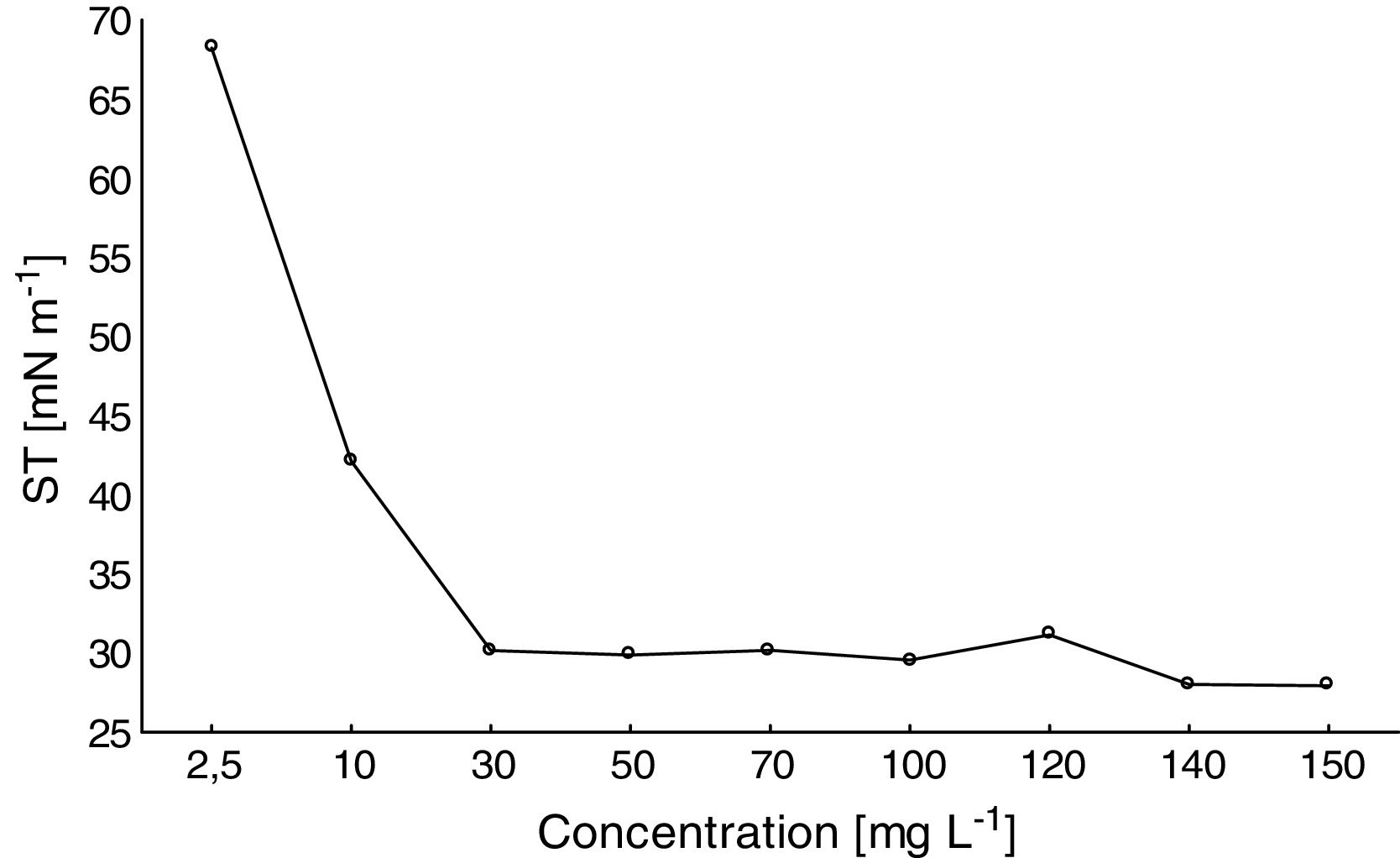
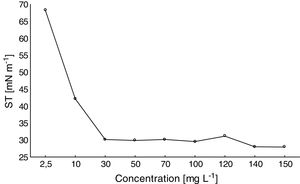
Determination of the CMCA satisfactory reduction of the ST of distilled water was observed with increasing amounts of the biosurfactant (Fig. 4). For biosurfactant concentrations above 30mgL−1, the ST remained stable at approximately 27mNm−1. Thus, the CMC of the biosurfactant produced by F. fujikuroi UFSM-BAS-01 was ∼30mgL−1.
Biosurfactant extraction, purification and identificationThe solvent system formed by ethyl acetate and methanol provided a satisfactory extraction of the biosurfactant produced by F. fujikuroi UFSM-BAS-01. The biosurfactant was extracted three times, and the second fraction yielded a compound in the form of large white crystals characteristic of sugars, from which substance C2, identified as the disaccharide α,β-trehalose, was obtained.20 Thus, the present study is the first report of the biosynthesis of an α,β-trehalose by F. fujikuroi, suggesting that the biosurfactant produced belongs to the class of trehalolipids.
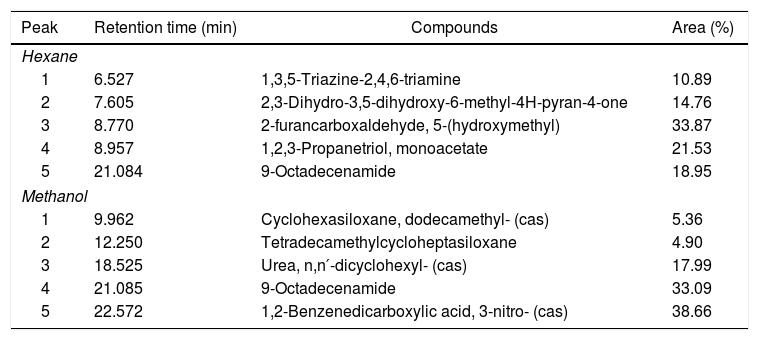
Compounds identified by GC–MS for both methanol and hexane extracts demonstrate the presence of 9-octadecenamide (oleamide), an amide derived from oleic acid, a fatty acid (Table 3). In GC-FID analysis, the methanol extract did not show evidence of any molecule with a fatty acid structure when compared with the FAME mix standard chromatogram pattern. However, the hexane extract exhibited a few molecules that can be correlated with the fatty acid pattern showed in the FAME mix standard chromatogram, such as caproic acid (41.72%) and palmitoleic acid (12.72%). Unfortunately, other peaks that would appear in the same chromatogram were not clarified due to the lack of derivatisation of the fatty acids, which is a subject for future work.
Volatile compounds of the hexane and methanol extracts of the biosurfactant produced by Fusarium fujikuroi UFSM-BAS-01 and analysed by GC–MS.
| Peak | Retention time (min) | Compounds | Area (%) |
|---|---|---|---|
| Hexane | |||
| 1 | 6.527 | 1,3,5-Triazine-2,4,6-triamine | 10.89 |
| 2 | 7.605 | 2,3-Dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one | 14.76 |
| 3 | 8.770 | 2-furancarboxaldehyde, 5-(hydroxymethyl) | 33.87 |
| 4 | 8.957 | 1,2,3-Propanetriol, monoacetate | 21.53 |
| 5 | 21.084 | 9-Octadecenamide | 18.95 |
| Methanol | |||
| 1 | 9.962 | Cyclohexasiloxane, dodecamethyl- (cas) | 5.36 |
| 2 | 12.250 | Tetradecamethylcycloheptasiloxane | 4.90 |
| 3 | 18.525 | Urea, n,n′-dicyclohexyl- (cas) | 17.99 |
| 4 | 21.085 | 9-Octadecenamide | 33.09 |
| 5 | 22.572 | 1,2-Benzenedicarboxylic acid, 3-nitro- (cas) | 38.66 |
The fungus UFSM-BAS-01 can be considered an efficient producer of biosurfactant. According to the literature, STs in the range of 35–40mNm−1 indicate a microorganism that is promising for biosurfactant production; values below 35mNm−1 indicate that the microorganism can be considered an efficient biosurfactant producer.21 The surface tension of the biosurfactant produced by the fungus UFSM-BAS-01, after optimization of the culture medium (20.08mNm−1), is lower than those found for Fusarium sp.22 and F. proliferatum,5 which were 32 and 36.6mNm−1, respectively.
Isolate UFSM-BAS-01 was identified by molecular techniques as the fungus F. fujikuroi (Giberella fujikuroi). This fungus belongs to the species complex F. fujikuroi, formerly named the G. fujikuroi complex, which is a monophyletic group that largely corresponds to the outdated Liseola section but also accommodates species originally classified in other Fusarium sections.23F. fujikuroi currently contains more than fifty species, which can be distinguished only by molecular parameters.8
The environmental variables that most influenced the production of the biosurfactant were temperature and incubation time. Several studies have demonstrated the positive influence of temperatures above 30°C on the production of biosurfactants by microorganisms.24–26 In our study, the biosurfactant with the lowest surface tension was produced with fermentation temperatures above 37°C, although the fungus was isolated and initially cultivated at a temperature of 30°C. Several authors cite 30°C as the ideal temperature for the production of biosurfactants by fungal species.22,25 However, the production of biosurfactants at high temperatures, at which microbial metabolism is accelerated, may facilitate the use of these molecules on an industrial scale, including studies on biosurfactants produced from thermophilic microorganisms.
Higher incubation times resulted in higher biosurfactant yields by the fungus F. fujikuroi UFSM-BAS-01. Similar results were observed by El-Sheshtawy et al.27 and Elazzazy et al.28 in the production of a biosurfactant by Bacillus licheniformis and Virgibacillus salarius, respectively. A significant reduction of the biosurfactant tension produced by F. fujikuroi (20.08mNm−1) was observed after seven days of incubation. Biosurfactants are secondary metabolites that are normally produced during stationary phases, which is likely why the greatest reduction of surface tension occurred only after seven days of fungal growth. The production of biosurfactants can occur or be stimulated by cell growth under limiting conditions.26
The agitation variable was statistically significant and had a negative influence on the production of the biosurfactant according to the Plackett–Burman design but was not significant in the central rotational compound design (CCRD). This was probably because the high agitation rates necessary to provide sufficient amounts of oxygen for cultures promote excessive foaming. This intense foaming decreases the yield of the process, as it removes part of the biomass and the substrate from the reaction medium, making it difficult to control the process.29
The ST and critical micellar concentration of the biosurfactant produced by F. fujikuroi UFSM-BAS-01 were similar to those of a biosurfactant produced by Bacillus subtilis YB7, which is capable of reducing the ST of distilled water from 70 to 30mNm−1and has a CMC 40mgL−1.30 In a similar study, Vaz et al.26 found a CMC of 40mgL−1 for a biosurfactant produced by B. subtilis EG1. The CMC of the biosurfactant produced by F. fujikuroi UFSM-BAS-01 is similar to that of the synthetic surfactant Findet® 1214N/23 (21mgL−1).31 This indicates that the biosurfactant produced by this fungus has the potential for commercial use and that this fungus could be used for the production of biosurfactants on an industrial scale.
The use of a solvent system containing ethyl acetate and methanol enabled the extraction of a disaccharide identified as trehalose.20 Until recently, the only form of trehalose that occurred naturally was α-d-glucopyranosyl-(1→1)-α-d-glucopyranoside, with three possible isomers (α,α-; α,β-; and β,β-). In our study, the fungus F. fujikuroi UFSM-BAS-01 produced an α,β-trehalose (neo-trehalose), which was recently biosynthesized by other fungi.32 Thus, the present study is the first report of the biosynthesis of an α,β-trehalose by F. fujikuroi, suggesting that the biosurfactant produced belongs to the class of trehalolipids. Different types of trehalose-containing glycolipids belonging to the group of mycolates are known to be produced by bacteria such as Mycobacterium, Rhodococcus, Arthrobacter, Nocardia and Gordonia.33 Trehalolipids have attracted interest for their potential applications in several areas due to their ability to decrease interfacial tension and increase the pseudosolubility of hydrophobic compounds.34
GC–MS analysis of methanol and hexane extracts of the biosurfactant produced by F. fujikuroi identified the presence of the lipophilic compound 9-octadecenamide, an oleamide. This lipophilic compound has many biological activities.35 Premjanu and Jaynthy36 found oleamide to be the major compound in an ethyl acetate extract of Colletotricum gloeosporioides fungal biomass. GC-FID hexane extract analysis showed several molecules that can be correlated with a fatty acid pattern, such as caprioic acid. However, additional analyses such as MALDI-TOF and FITR are needed to confirm further details about the structure of the biosurfactant.
Conclusion- -
The fungus F. fujikuroi UFSM-BAS-01 isolated from soil samples contaminated with hydrocarbons is an efficient biosurfactant producer;
- -
The production of biosurfactants by F. fujikuroi UFSM-BAS-01 is significantly increased under optimized culture conditions;
- -
The preliminary identification of the structure of the biosurfactant demonstrates the presence of an α,β-trehalose, suggesting that the biosurfactant belongs to the class of trehalolipids.
The authors declare no conflicts of interest.
The authors thank the Foundation for Research Support of the State of Rio Grande do Sul (FAPERGS), National Council of Technological and Scientific Development (CNPq) and Coordination for the Improvement of Higher Level or Education Personnel (CAPES) for providing scholarships and financial support of this work.