Metallothioneins are a superfamily of low-molecular-weight, cysteine (Cys)-rich proteins that are believed to play important roles in protection against metal toxicity and oxidative stress. The main purpose of this study was to investigate the effect of heterologous expression of a rice metallothionein isoform (OsMTI-1b) on the tolerance of Saccharomyces cerevisiae to Cd2+, H2O2 and ethanol stress. The gene encoding OsMTI-1b was cloned into p426GPD as a yeast expression vector. The new construct was transformed to competent cells of S. cerevisiae. After verification of heterologous expression of OsMTI-1b, the new strain and control were grown under stress conditions. In comparison to control strain, the transformed S. cerevisiae cells expressing OsMTI-1b showed more tolerance to Cd2+ and accumulated more Cd2+ ions when they were grown in the medium containing CdCl2. In addition, the heterologous expression of GST-OsMTI-1b conferred H2O2 and ethanol tolerance to S. cerevisiae cells. The results indicate that heterologous expression of plant MT isoforms can enhance the tolerance of S. cerevisiae to multiple stresses.
The yeast, Saccharomyces cerevisiae, has several properties which have established it as an important tool in the expression of foreign proteins for research, industrial or medical use.1,2 As a food organism, it is highly acceptable for the production of pharmaceutical proteins. In contrast, Escherichia coli has toxic cell wall pyrogens and mammalian cells may contain oncogenic or viral DNA, so that products from these organisms must be tested more extensively.3,4S. cerevisiae can be grown rapidly on simple media and to high cell density, its genetics are more advanced than any other eukaryote, and can be manipulated almost as readily as E. coli. As a eukaryote, S. cerevisiae is a suitable host organism for the high-level production of secreted as well as soluble cytosolic proteins.5
Industrial yeast strains during fermentation are exposed to various stresses such as osmotic shock, oxidative stress and toxicity of secondary metabolites which lead to the loss of biological products.6,7 Molecular oxygen is relatively unreactive and harmless in its ground state, but can undergo partial reduction to form a number of reactive oxygen species (ROS), including the superoxide anion and hydrogen peroxide (H2O2), which can further react to produce the highly reactive hydroxyl radical.8 ROS are toxic agents that can damage a wide variety of cellular components resulting in lipid peroxidation, protein oxidation, and genetic damage through the DNA modification.8,9 Yeast cells may be exposed to ROS generated by neutrophils and macrophages during immunological defense mechanisms and following exposure to numerous exogenous agents including xenobiotics, carcinogens, UV and ionizing radiation.7
S. cerevisiae, like all organisms, contains effective antioxidant defense mechanisms, which detoxify ROS as they are generated and maintain the intracellular redox environment in a reduced state.9 In response to an oxidant challenge, yeast cells increase the synthesis of a number of antioxidant enzymes involved in the detoxification of ROS including glutathione reductase, superoxide dismutase, catalase, glutathione peroxidase and thioredoxins.9,10 In addition to antioxidant enzymes, growing amount of evidences show a close relationship between oxidative stress and metallothioneins (MTs); the low molecular weight Cys-rich proteins which are ubiquitously present in eukaryotes and prokaryotes. In fact a variety of physical and chemical stresses associated with oxidative injury increase MTs synthesis.11–13
MTs belong to a superfamily of intracellular metal-binding proteins which is composed of 15 families.14,15 MTs bind metals through the thiol groups of their Cys residues.14,16 Induction of MTs synthesis by heavy metals is a very well-known phenomenon and accordingly MTs are regarded as biomarker to reflect heavy metal exposures in ecotoxicological assessment.15,17 MTs have a strong effect in scavenging free radicals, which are produced under various stress conditions.13 In Arabidopsis, it has been demonstrated that different types of MTs exhibit distinct and overlapping functions in maintaining the homoeostasis of essential transition metals, detoxification of toxic metals, and protection against intracellular oxidative stresses.18–21 Transgenic tobacco and yeast that overexpress MT from cotton displayed increased tolerance to environmental stresses, indicating the role of MT in response to abiotic stresses.22
Previously we heterologously expressed the isoform OsMTI-1b, a rice MT type 1, in E. coli as carboxyl-terminal extensions of glutathione-S-transferase (GST).15,23 The cells expressing OsMTI-1b showed increased tolerance to Ni2+, Cd2+, and Zn2+ and accumulated more metal ions compared with cells expressing GST alone. In addition, heterologous expression of OsMTI-1b conferred H2O2 tolerance to E. coli cells.15
In this study, the gene encoding OsMTI-1b was heterologously expressed in S. cerevisiae. The tolerance of transgenic S. cerevisiae to cadmium, hydrogen peroxide as well as ethanol was examined and the accumulation of cadmium in the cells was determined.
Materials and methodsCloning of genes encoding GST-OsMTI-1bIn order to clone the gene encoding GST-OsMTI-1b, the plasmid pET41a-OsMTI-1b was used as template.15 The DNA fragment containing GST-OsMTI-1b was amplified using Pfu DNA polymerase (Thermo Scientific) in a reaction mixture containing template plasmid, deoxy nucleotides, reaction buffer and the primers 5′-ATATAAGCTTG CGGATAACAATTCCCCTCT-3′, which carries an HindIII restriction site at the 5′ end (underlined) of forward primer, and 5′-ATTTCTCGAGTTAGCAGTTGCAAGGGTT-3′ with a restriction site XhoI (underlined) at the 5′ end of reverse primer. An addition of four bases was included at the 5′ end in each oligonucleotide primer. The thermal profile was as follows: 1 cycle at 92°C for 5min; 35 cycles at 92°C for 1min; 63°C for 1.5min; 72°C for 2min and 1 cycle at 72°C for 10min. The PCR product was then digested with enzymes HindIII and XhoI and ligated using T4 ligase into p426GPD as a yeast expression vector,24 after linearization with HindIII and XhoI. The ligation mix was used to transform competent E. coli cells (DH5α). Transformants were selected on LB medium supplemented with ampicillin (50mgL−1). The resulting plasmid was termed GPD-GST-OsMTI-1b. The resulting plasmids were then verified by sequencing.
Heterologous expression of OsMTI-1b in S. cerevisaeThe competent cells from S. cerevisiae strain CEN.PK 113-5D (5D) were provided by using lithium acetate method as previously described.25 The new constructs GPD-GST-OsMTI-1b was transformed to competent cells of 5D to give new strain 5D-2527. A control strain was provided by transforming empty GPD to competent cells. Transformants were selected on plates containing SC-URA medium after incubation at 30°C for 2–3 days.
The new strain 5D-2527 and control were grown in shake flasks containing 10mL appropriate minimal medium [10% (NH4)2SO4, 12% KH2PO4, 5% MgSO4, trace metals 1mLL−1, 2% glucose and vitamins] at 30°C under vigorous agitation for overnight. Then 4mL of the overnight cultures of cells were inoculated in 80mL of minimal medium supplemented with 2% glucose. When the O.D.600 of cultures were about 2.5 and 3.5, 1mL samples of culture medium were harvested by centrifugation at 12,000×g for 15min, and frozen at −20°C until use.
Extraction and identification of recombinant proteins with western blot analysesThe extraction of total protein was performed as explained previously.26 For extraction of total proteins, the frozen pellets were first pre-treated with 2.0M LiAc for 10min at room temperature. After centrifugation (12,000×g for 10min, 4°C), the remaining pellets were resuspended in 0.4M NaOH for 5min on ice, followed by centrifuging (12,000×g for 10min, 4°C), the remaining pellets were resuspended in 100μL SDS sample buffer (0.06M Tris–HC1 pH 6.8, 10% (v/v) glycerol, 2% (w/v) sodium dodecyl sulfate (SDS), 5% (v/v) 2-mercaptoethanol, 0.0025% (w/v) bromophenol blue) and boiled for 5min. Then, the cell lysate was centrifuged to remove the cell debris. The total proteins recovered in supernatant phase were analyzed by 12% SDS-PAGE and stained by Coomasie Brilliant Blue R-250.27
For western blotting, proteins were transferred to a PVDF membrane (Roche Applied Science) according to the manufacturer's instructions. The blots were blocked overnight in blocking buffer containing 5% (w/v) skimmed milk in TBST (10mmolL−1 NaCl, 25mmolL−1 Tris–HCl, pH 7.5, 0.1% (v/v) Tween 20). The rabbit His-Tag antibody (GenScript), was diluted 1:1000 in TBST. The antigen–antibody interaction was carried out at room temperature for 1h. The blots were washed (3× 10min) with TBST followed by (1× 10min) TBS (10mmolL−1 NaCl, 25mmolL−1 Tris–HCl, pH 7.5). Blots were then probed with goat-anti-rabbit IgG conjugated with horse radish peroxidase (GenScript) diluted 1:2000 in TBST as secondary antibody. The membranes were washed again, as explained before. After washing, the immune blots were developed using 0.5mgmL−1 diaminobenzidine in 50mmolL−1 Tris–HCl, pH 7 and 0.22% hydrogen peroxide.
Tolerance of S. cerevisiae cells to Cd2+, H2O2 and ethanolTolerance of new strain and control to metals, H2O2 and ethanol in the growth medium was examined in different concentrations of CdCl2·H2O (0.3 and 0.9mM), H2O2 (1 and 3mM) and ethanol (7 (v/v) % and 10 (v/v) %). For these analyses, 5mL samples of the overnight cultures were inoculated in 80mL of minimal medium supplemented with 2% glucose. The cultures were incubated at 30°C with shaking. The culture media containing 5D-2527 or control strain were supplemented with cadmium at concentrations of 0.3 and 0.9mM when O.D.600 reached 3.3. The growth was monitored up to 14h by O.D.600 measurements. Each data represents the mean±SD obtained from three independent experiments with two replicates. For the analysis of cadmium in medium, cells from 10mL of culture at 0 (T0) and 5h after cadmium addition (T1) were precipitated by centrifugation at 6000×g for 20min. The supernatant was analyzed for Cd2+ using inductively coupled plasma atomic absorption spectroscopy (Perkin Elmer AAnalyst 700). The cadmium concentration changes in the medium of control and 5D-2527 between T1 and T0 (CT0–CT1) were calculated. Each data is the mean±SD obtained from three independent experiments with two replicates. Differences at the 5% level were considered significant as analyzed by the paired student's t-test (p<0.05).
For analysis of H2O2 and ethanol tolerance, H2O2 and ethanol were added to the culture media after 7h when O.D.600 reached 3.6. The growth was monitored up to 14h by O.D.600 measurements. Each data represents the mean±SD obtained from three independent experiments with two replicates. Differences at the 5% level were considered significant as analyzed by the paired student's t-test (p<0.05).
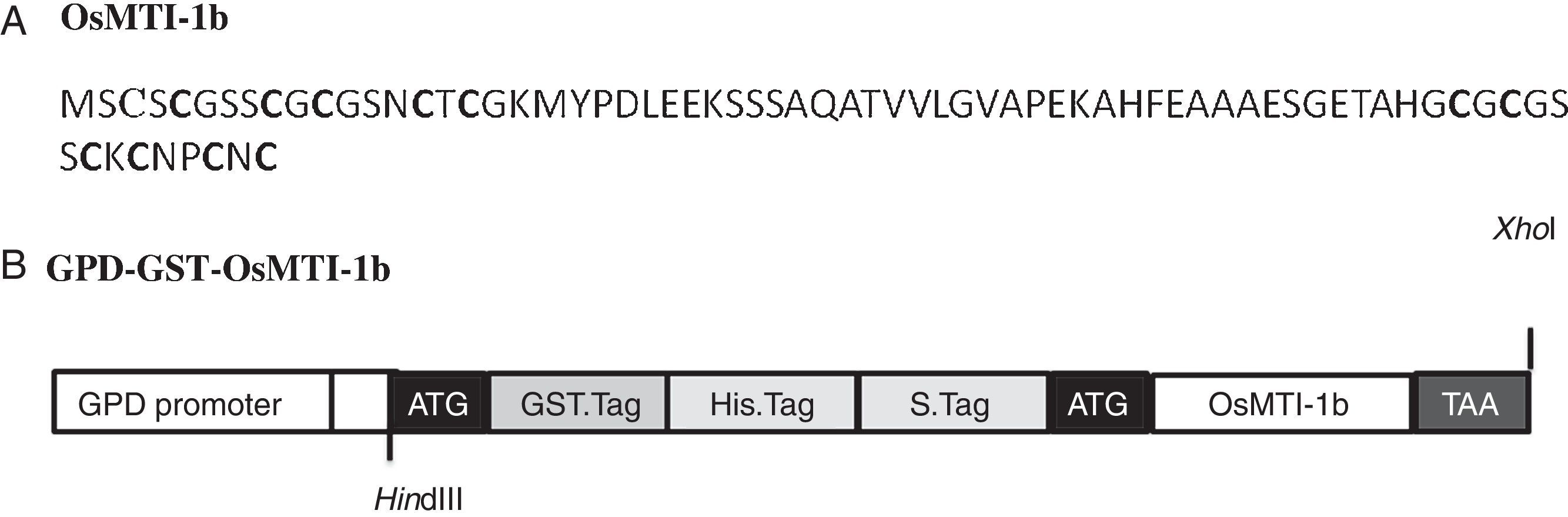
ResultsHeterologous expression of GST-OsMTI-1b in S. cerevisiaeThe full-length coding sequence of OsMTI-1b consists of 219bp encoding a protein with 72 amino acids and theoretical molecular weight (kDa)/pI of 7.13/5.08. The predicted protein consists of six C–X–C motifs equally distributed in N- and C-terminals, as other plant type 1 MT proteins (Fig. 1A). The protein was expressed as carboxyl-terminal extensions of GST.tag, 6 His.tag and S.tag which we entirely named GST in this study (Fig. 1B). The use of GST has the advantages of high solubility and stability against proteolytic degradation.28 Therefore, the protein GST-OsMTI-1b is predicted to have theoretical molecular weight (kDa)/pI of 39.94/5.93. Western blot analysis showed a sharp protein band of expected molecular mass (Fig. 2, lanes 3 and 4).
SDS-PAGE and western blot analysis of total proteins extracted from control strain (lanes 1 and 2) and 5D-2527 (lanes 3 and 4) when the O.D.600 of cell cultures were 2.5 (lanes 1 and 3) and 3.5 (lanes 2 and 4). The purified GST-OsMTI-1b obtained from heterologous expression in E. coli15 was run in lane P.
The comparison of growth curves of strain 5D-2527 and control reveals that in the H2O2/ethanol free medium, the maximum O.D.600 for control strain (O.D.max600 6.6±0.2) is almost higher than that for the strain 5D-2527 (O.D.max600 6±0.2). However, in the presence of 1mM H2O2 the maximum O.D.600 of strain 5D-2527 (O.D.max 6.2±0.24) was higher than that for the control strain (O.D.max600 4.6±0.2, Fig. 3 A). In the presence of 3mM H2O2 the maximum O.D.600 of strains 5D-2527 and control strain were 4.8±0.2 and 3.8±0.17, respectively. In addition, in the presence of 7 and 10% ethanol the maximum O.D.600 values for the control strain were 4.6±0.2 and 4.2±0.2, respectively. These values were reached 5.6±0.18 and 5.1±0.25 for 5D-2527 in the same conditions (Fig. 3B). These results suggest that the heterologous expression of GST-OsMTI-1b confers H2O2 and ethanol tolerance in S. cerevisiae cells.
Effects of heterologous expression of GST-OsMTI-1b on the tolerance of S. cerevisiae to (A) H2O2 (1 and 3mM), (B) ethanol (7% and 10%) and (C) Cd2+ (0.3 and 0.9mM). Each data is the mean±SD obtained from three independent experiments with two replicates. Differences at the 5% level were considered significant as analyzed by the paired student's t-test (p<0.05).
In the presence of 0.3 and 0.9mM Cd2+ the maximum O.D.600 for control strain were 3.2±0.25 and 3.4±0.2, respectively. These values reached 4±0.2 and 4.7±0.15 for the strain 5D-2527 when it was grown in the same conditions. These results suggest that the heterologous expression of GST-OsMTI-1b confers cadmium tolerance in S. cerevisiae cells.
To determine whether the enhanced of tolerance of 5D-2527 to cadmium was due to increased accumulation of this metal ion in the cells, the concentrations of cadmium ions in the culture media inoculated with the 5D-2527 and control strains were determined by atomic absorption at 0h (T0) and 5h (T1) after the addition of cadmium to the cultures. The concentration of cadmium ions at T1 was similar to that at T0 for the control strain whereas the concentrations of cadmium ions were decreased by 5.57ppm at T1 in the culture medium of 5D-2527 (Fig. 4).
Cadmium concentration variation in the medium of control strain and strain 5D-2527 between T1 (5h after addition CdCl2 to medium) and T0 (starting point of addition CdCl2). Each data is the mean±SD obtained from three independent experiments with two replicates. Asterisks indicate statistically significant differences from the corresponding control as analyzed by the paired student's t-test (p<0.05).
S. cerevisiae is the most important industrial yeast and the main microorganism employed in bioethanol production.29 Industrial yeast strains during fermentation are exposed to various stresses such as osmotic shock, oxidative stress and toxicity of secondary metabolites which lead to the loss of biological products.6,29 During oxidative stress in S. cerevisiae the protein Yap1p as a transcription factor plays a key role in activation of transcription of many genes including TRX2,30 encoding thioredoxin, TRR1 – thioredoxin reductase,31GSH2 – glutathione synthase, GPX2 – glutathione peroxidase 232 and TSA1 – thioredoxin peroxidase133,34 via binding with specific DNA sequences localized in their promoters.35 Correspondingly the over expression of genes TRX2 and TRR1 enhanced the resistance of S. cerevisiae to oxidative stress.30,31
Ethanol production by S. cerevisiae is one of the first biotechnological commodities for many years. However the inhibitory effect of ethanol accumulation on yeast growth during fermentation is still an important challenge for bio-ethanol production.36 Depending on yeast strain and condition, ethanol inhibition on yeast growth begins at concentrations of less than 5%.37 The mechanisms that yeasts cope with ethanol stress are very complicated and not fully understood. The quantification of the growth behavior of all available single gene deletion strains of S. cerevisiae under ethanol stress revealed that the growth of 446 deletion strains was defective indicating that many genes are involved in the ethanol resistance in yeast. The deletion of some of these genes like PEX genes whose products are involved in peroxisome transport exhibited the severest sensitivity to ethanol.38 Recently the generation of new transgenic S. cerevisiae strains has been successfully implemented to improve ethanol tolerance.39–42 For instance, the overexpression of genes INO1, DOG1, HAL1 and truncated form of MSN2 in S. cerevisiae resulted in remarkably increased tolerance to high concentrations of ethanol.41
In the present work the gene encoding OsMTI-1b was transferred to S. cerevisiae. MTs are Cys-rich and low molecular weight proteins characterized as important intracellular factors in detoxification of heavy metals in the cells of prokaryotes and eukaryotes.14,43 However the role of MTs in the protection of plant and animal cells against oxidative stress has been recently demonstrated.22,44,45 The isoform OsMTI-1b is known as a rice MT type 1 due to the presence of six Cys residues in N- and C-terminals. Previously this isoform was heterologously expressed in E. coli. Transformed cells showed increased tolerance to heavy metals cadmium, zinc and nickel and accumulated more metals as compared to the control strain.15,23 Furthermore, the heterologous expression of OsMTI-1b conferred hydrogen peroxide tolerance to E. coli cells.15 The data presented here show that the heterologous expression of OsMTI-1b was successful when it was expressed as carboxy-terminal extension of GST. The S. cerevisiae cells expressing GST-OsMTI-1b were able to remove efficiently cadmium from culture medium and showed more tolerance in the medium containing cadmium as compared to the control strain. In addition, the cells expressing GST-OsMTI-1b conferred H2O2 tolerance to S. cerevisiae. Similar results have been previously reported for transgenic yeasts overexpressing cotton MT type 3 (GhMT3) in response to H2O2 stress22 or MT type 3 from Tamarix hispida (Th MT3) in response to metal toxicity.45 Here we also found that the expression of GST-OsMTI-1b enhances the resistance of transgenic S. cerevisiae against ethanol.
Taken together on the basis of our results, the heterologous expression of isoform OsMTI-1b conferred improved tolerance against H2O2, metals and ethanol. Therefore plant genes encoding MT isoforms may be useful for augmenting the genetic toolbox for generating new strains with multiple stress resistance which could be very efficient for biofuel production.
Conflicts of interestThe authors declare that there is no conflict of interest regarding the publication of this paper.
Iran National Science Foundation is thanked for financial support (grant No. 91001342).