Metabolites of mycoparasitic fungal species such as Trichoderma harzianum 88 have important biological roles. In this study, two new ketoacyl synthase (KS) fragments were isolated from cultured Trichoderma harzianum 88 mycelia using degenerate primers and analysed using a phylogenetic tree. The gene fragments were determined to be present as single copies in Trichoderma harzianum 88 through southern blot analysis using digoxigenin-labelled KS gene fragments as probes. The complete sequence analysis in formation of pksT-1 (5669bp) and pksT-2 (7901bp) suggests that pksT-1 exhibited features of a non-reducing type I fungal PKS, whereas pksT-2 exhibited features of a highly reducing type I fungal PKS. Reverse transcription polymerase chain reaction indicated that the isolated genes are differentially regulated in Trichoderma harzianum 88 during challenge with three fungal plant pathogens, which suggests that they participate in the response of Trichoderma harzianum 88 to fungal plant pathogens. Furthermore, disruption of the pksT-2 encoding ketosynthase–acyltransferase domains through Agrobacterium-mediated gene transformation indicated that pksT-2 is a key factor for conidial pigmentation in Trichoderma harzianum 88.
Biological control is an efficient and environmentally friendly way to prevent damping off. Trichoderma spp. are opportunistic biological control agents and avirulent plant symbionts. T. harzianum belonging to Trichoderma spp. is widely used as a biopesticide and biofertilizer to control diseases and promote positive physiologic responses in plants. It combats several plant pathogenic fungi such as Fusarium oxysporum in bean and melon,1,2Botrytis cinerea in tobacco,3Rhizoctonia solani in cucumber,4 sheath blight in rice,5 creeping bentgrass and tomato and Sclerotinia sclerotiorum in soybean.6–8 These reports suggest that T. harzianum strains parasitize a large variety of phytopathogenic fungi. It can also be used to control R. solani.9 Therefore, the molecular biocontrol mechanism of T. harzianum deserves further study. T. harzianum is reportedly effective in controlling fungi through different mechanisms, such as by inducing systemic resistance in plants, parasitizing other fungi by recognizing signals from the host to provoke the transcription of mycoparasitism-related genes, enzyme production and secretion, competing for nutrients and light and by producing antifungal metabolites.10 Harman et al.11 reported that T. harzianum strains significantly enhance the systemic resistance and seed growth of maize, and induces the expression of proteins related to systemic induced resistance and increases growth.12 Furthermore, although the underlying mechanism is much more complex than previously considered, the symbiosis-like interaction can be genetically improved to benefit plants more.13 Proteins such as G-proteins and mitogen-activated protein kinases affect biocontrol-relevant processes in the mycoparasitic interactions between Trichoderma and plant pathogens, such as the production of hydrolytic enzymes and antifungal metabolites and the formation of infection structures.14 The transcriptional factor Pac1 regulates some proteins involved in antagonism, such as chit42 chitinase, papA protease, gtt1 glucose permease, and qid74 cell wall protein.15 Competition for nutrients and space is also considered antagonism. T. harzianum showed dominance over Alternaria alternata under different environmental conditions (water activity and temperature).16 However, an in vitro antagonism test indicates that the production of antifungal compounds under nutrient-limiting conditions also contributes to fungistasis.17 In general, elucidating the pathogen-induced biocontrol mechanism of T. harzianum enables its application in controlling plant diseases. T. harzianum is currently used as a biofungicide to control F. oxysporum and Sclerotium rolfsii efficiently.18
Antifungal metabolites isolated from Trichoderma spp., including non-ribosomal peptides, polyketides, terpenoids and pyrones, play the important role in biological control.10 And studying biological activities, especially their biosynthetic pathway of those compounds is necessary for uncovering their biological control mechanisms.
Polyketide synthases (PKSs) are major enzymes in plants, fungi, and bacteria that are responsible for polyketide biosynthesis.19 Fungal PKSs are generally large multidomain systems that comprise a minimal set of ketosynthase, acyltransferase and acyl carrier protein domains. Type I PKSs elongate their polyketide products iteratively. In addition, ketoreductase, dehydratase and enoyl reductase domains may catalyse optional β-keto processing reactions, and optional accessory domains provide cyclase activity and methyl transferase activity.20 Type I fungal PKSs are grouped into three classes based on their optional processing domains: non-reducing (e.g., melanin, aflatoxin), partially reducing (e.g., 6msas, calicheamicin), and highly reducing PKSs (e.g., T-toxin, fumonisin).8,21
A variety of fungal polyketide synthase genes have been identified in recent ten years. However, there were no related research reports that polyketide synthase genes of T. harzianum were cloned and identified. In this study, the genes encoding type I PKSs from T. harzianum mycelia were isolated using gene walking technology, and predictive analysis of its structure and encoding product. And PKS genes expression from different growth stages of antagonism between T. harzianum and fungal pathogens was detected by real-time quantitative RT-PCR (RT-qPCR) method. Furthermore, PKS gene function was studied by gene knockout technology. These studies were foundation for further revealed PKS in the role of biological metabolic process and its biological activity, and also provide new research ideas for future study of other fungi PKS gene.
Materials and methodsFungal strainsThe biocontrol agent T. harzianum strain 88 (CGMCC 5403; GenBank accession no. of ITS: JN579713) and the fungal plant pathogens Rhizoctonia solani (ACCC 36246), S. sclerotiorum (ACCC 36462) and F. oxysporum (ACCC 36966) were stored at the Microbial Genetic Engineering Laboratory, Harbin Institute of Technology (Harbin, China).
Isolation of PKS genes via gene walking using degenerate primersTotal DNA was isolated from T. harzianum 88 using a DNeasy Plant Mini kit (Invitrogen, Carlsbad, CA, USA). PKS ketoacyl synthase (KS) and acyltransferase (AT) sequences were amplified using the ketosynthase–acyltransferase (KA) degenerate primers KAF1 and KAR1 previously designed and reported by Amnuaykanjanasin et al.22 The PCR cycling conditions were as follows: 94°C for 5min; 30 cycles of 94°C for 30s, 58°C for 30s and 72°C for 30s; and 72°C for 7min. The resulting 700bp product was gel isolated and inserted into the pMD18-T Easy vector (Promega, Madison, WI, USA). The recombinant vector was then transformed into competent Escherichia coli DH5α (TaKaRa, Osaka, Japan) using the heat shock method.23 The transformants were spread onto LB agar medium (1% tryptone, 0.5% yeast extract, 1% NaCl and 1.5% agar at pH 7.0) (TaKaRa, Osaka, Japan) containing ampicillin (100μg/mL) (TaKaRa, Osaka, Japan), X-Gal (0.2mM) (TaKaRa, Osaka, Japan) and isopropylthio-β-galactoside (40μg/mL) (TaKaRa, Osaka, Japan) and incubated overnight at 37°C. White colonies were selected for PCR identification.
The forward primer xKAF1 was designed based on the amino acid (aa) sequences in the conserved KS domain of reducing fungal PKS. The xKAF1 forward and KAR2 reverse primers were used to isolate an 800bp fragment, which was cloned into the pMD18-T Easy vector (Promega, Madison, WI, USA) and sequenced.
A set of sense primers for 3′ end amplification and antisense primers for 5′ end amplification were designed for gene walking to obtain more complete genomic sequences of the two PKS genes. The reverse primers AP1, AP2, AP3 and AP4 for the two PKS genes were synthesized by TaKaRa (Osaka, Japan). The sequences of all primers used in this study are listed in the Online Resource 1. The fragment sequences were assembled by Phrap (http://www.phrap.org/) and were then analysed using the BLASTx algorithm (http://www.ncbi.nlm.nih.gov/BLASTx/) and ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The putative PKSs were designated as pksT.
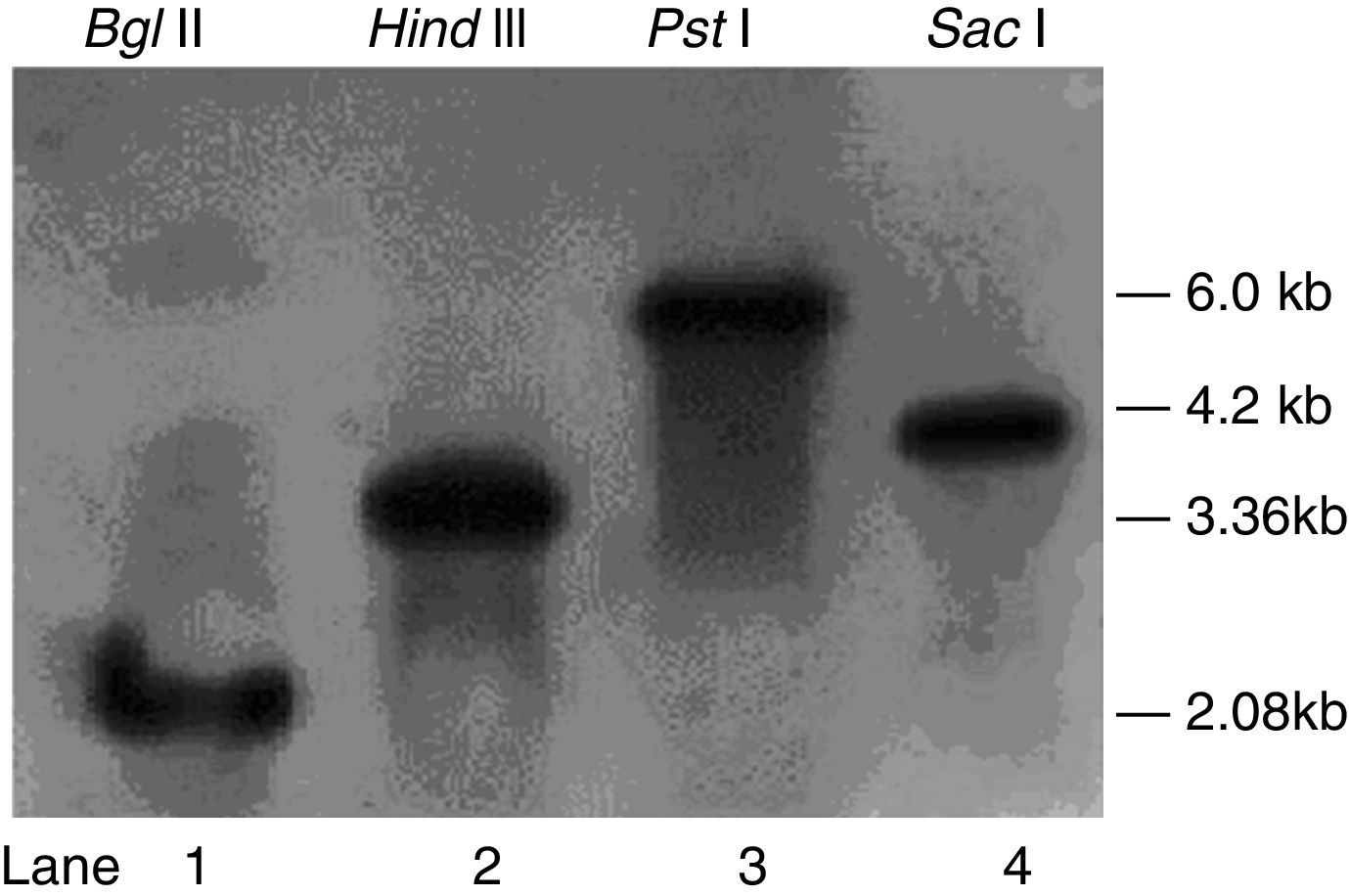
Southern blot analysisTwo specific primer pairs were designed according to the KS domain fragment sequences, as shown in the schematic provided in the Online Resource 1. The DIG-labelled DNA probes pksT-a-S and pksT-b-S were generated from the cloned KS domain PCR products using a DIG-High Prime Labelling Mix (Roche Applied Sciences, Basel, Switzerland). Approximately 180μg of genomic DNA was divided into six equal portions and digested overnight at 37°C with the restriction enzymes BglII (used to digest two portions), HindIII, SalI or SacI (Promega, Madison, WI, USA) to ensure complete digestion. The digested genomic DNA was then electrophoresis on 0.7% (w/v) agarose gel (30V, 12h) and transferred onto positively charged nylon membranes (GE Healthcare, London, UK). The membranes were hybridized at 42°C using the DIG-labelled probes pksT-a-S and pksT-b-S (sequences provided in Online Resource 2) and were detected with the DIG Luminescent Detection System (Roche Applied Sciences, Basel, Switzerland).
Reverse transcription-polymerase chain reaction (RT-PCR) of PKS genesT. harzianum 88 mycelia (100mg) were ground into powder after freezing in liquid nitrogen. RNA was then extracted using an RNeasy Plant Mini kit (Invitrogen, Carlsbad, CA, USA) and used for first-strand cDNA synthesis with SuperScriptII reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The full cDNA sequence of T. harzianum 88 pksT was amplified using RT-PCR primers specific for pksT-1 and pksT-2 (Online Resource 1). The amplified fragments were then assembled using the program Phrap (http://www.phrap.org/).
The potential open reading frames (ORFs) of the two PKSs were scanned using FGENESH as the matrix (http://linux1.softberry.com) and compared with T. virens gene sequences. The conserved PKS domains were verified using CDD Search/PRS-BLAST (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The pksT KS domains were aligned with 55 fungal PKS protein sequences (reducing and non-reducing) retrieved from GenBank, and the two alignments analysed using CLUSTALW were embedded in the MEGA 4.0 program (http://www.megasoftware.net/). Bootstrap analysis was conducted using the MEGA 4.0 program with 1000 replications to obtain the confidence value for the aligned sequence dataset. A phylogenetic tree was constructed via maximum parsimony. The resultant phylogenetic tree was rooted in animal fatty acid synthase (Gallus gallus FAS), as previously described.21 The candidate secondary metabolite product was predicted using Natural Product Domain Seeker (NaPDoS) (http://npdomainseeker.sdsc.edu//run_analysis.html).
Differential expression of PKS genesRT-qPCR was used to evaluate the relative changes in gene expression in T. harzianum 88 during challenge with the fungal pathogens R. solani, S. sclerotiorum and F. oxysporum. T. harzianum 88 was challenged with R. solani, S. sclerotiorum and F. oxysporum on plates using the procedure described by Carsolio et al.24 Agar with cut fungal colonies were placed on opposite sides of 9cm plates containing minimum medium (MM) with 2% agar and 2% glucose (Promega, Madison, WI, USA) and were covered with sterile cellophane sheets (Promega, Madison, WI, USA). T. harzianum 88 mycelia were collected when they touched the fungal pathogen at 3, 6, 12, 24, and 36h. The procedure was repeated three times; however, only Trichoderma mycelia without spores and 0.5cm away from the original inocula were collected. The control plate was T. harzianum 88 challenged with T. harzianum 88 mycelia, which were collected when the colonies touched each other (3, 6, 12, 24, and 36h). The fungi were allowed to grow at 28°C in the dark. Two primer pairs were selected for real-time RT-PCR analysis (Online Resource 1).
Total RNA was extracted from the mycelia and digested with DNase I (TaKaRa, Osaka, Japan).25 Total RNA (4μg) from each pooled sample was reverse transcribed into cDNA in the presence of oligo (dT) in a total volume of 25μL. The synthesized cDNA was diluted with 25μL of RNase-free ddH2O and used as the template for RT-qPCR. Three pairs of primers were designed according to the ITS DNA from R. solani, S. sclerotiorum, and F. oxysporum to avoid the amplification of host fragments. Consequently, no fragments were amplified from cDNA templates. β-Tubulin (DY762540) transcripts were used as endogenous references for RT-qPCR. The primer set for β-tubulin was designed (additional data are given in the Online Resource 1) and RT-qPCR was performed using the SYBR® Premix Ex Taq™ II (TaKaRa, Osaka, Japan) on an ABI Model 7500 HT sequence detector (ABI, Harbin Institute of Technology, China). Reagent mixes were prepared by combining 12.5mL of SYBR® Premix Ex Taq™ II (2×) with 2.5μL of cDNA, 0.4pmol/μL forward primers, 0.4pmol/μL reverse primers, and 8μL distilled water per reaction. The 25μL reactions were carried out using programmed thermal cycling conditions consisting of 30s at 95°C, followed by 40 cycles of 5s at 95°C and 40s at the appropriate annealing temperature for each real-time RT-PCR primer set. Three technical replicates of each of the three biological replicates were used to normalize the relative quantification analysis. Data from the threshold cycles were analysed according to the 2 (ΔΔC(T)) methods in PKS gene expression.26
A Student's t-test was used to determine whether the pksT genes significantly affected the survival of T. harzianum 88 with and without challenge from the three fungal plant pathogens. The level of significance of Student's t-test was set to p<0.05 and was noted at p<0.01.
Determination of optimal hygromycin B concentrationThe wild type isolate (T) was assayed for growth on hygromycin B (hyg) (Promega, Madison, WI, USA) to optimize the negative selection for subsequent transformations. Agar pieces from plate cultures of the isolate were used to inoculate M-100 plates amended with varying hyg concentrations. Individual wells were photographed after 3 days.8
Construction of pksT-2 disruptant strainStandard molecular techniques were used to construct all plasmids using restriction enzymes (Promega, Madison, WI, USA), alkaline phosphatase and PCR reagents (TaKaRa Bio Inc., Otsu, Japan). The plasmids were purified using the MiniBEST Plasmid Purification Kit Ver.3.0 (QIAgen, USA). The binary vector pPZPtk8.10 was from Gardiner and Howlett.27
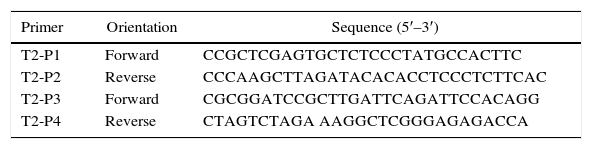
The 5′-flanking DNA (1.4kb, 5′ and 3′) and 3′-flanking DNA (1.114kb, 5′ and 3′) of pksT-2 were amplified by PCR using T. harzianum 88 genomic DNA as the template. The primers used to amplify the 5′-flanking DNA were T2-P1 and T2-P2, whereas those for the 3′-flanking DNA were T2-P3 and T2-P4 (Table 1). The 3′-flanking DNA was digested using BamHI and XbaI (Promega, Madison, WI, USA), and the digested fragment was ligated into the BamHI and XbaI site of the pPZPtk8.10. The generated plasmid was designated as pPZPtk8.10-ΔLR. The 5′-flanking DNA was digested with XhoI and HindIII (Promega, Madison, WI, USA), and the digested fragment was ligated into the XhoI and HindIII (Promega, Madison, WI, USA) site of the pPZPtk8.10. The generated plasmid was designated as pPZPtk8.10-ΔRR-ΔLR. The hygromycin resistance gene Hyg was amplified via PCR using the plasmid pCSN43 (Fungal Genetics Stock Center, Kansas, USA) as a template and primers hpg cassette F and hpg cassette R (Online Resource 1). The DNA fragment and pPZPtk8.10-ΔRR-ΔLR were digested using HpaI (Promega, Madison, WI, USA), and the digested fragment was ligated into the blunt-end restriction HpaI site of the pPZPtk8.10-ΔRR-ΔLR vector, which was treated with alkaline phosphatase. The generated plasmid was designated as pPZPtk8.10-ΔpksT-2.
Fungal transformationA. tumefaciens-mediated transformation of T. harzianum 88 was performed as described for other fungi with minor modifications. Briefly, A. tumefaciens carrying the plasmid DNA was grown on solid LB medium containing 50μg/mL kanamycin and 50μg/mL streptomycin (LB-ka-str) (Promega, Madison, WI, USA) at 28°C for 24h. Single colonies were inoculated into liquid (LB-ka-str) and incubated at 28°C for 12h. The bacterial culture was liquid induction medium (IM) (10mM K2HPO4, 10mM KH2PO4, 2.5mM NaCl, 2mM MgSO4, 0.7mM CaCl2, 9nM FeSO4, 4mM NH4SO4, 10mM glucose, 40mM 2-[N-morpholino] ethanesulfonic acid, pH 5.3, and 0.5% glycerol) containing 200μM acetosyringone (4′-hydroxy-3′,5′-dimethoxyacetophenone) AS (Promega, Madison, WI, USA) to an optical density of 0.15at 660nm.8 After cultivation of bacterial cells at 28°C for 6h, 1×106 fungal conidia/mL was mixed with the bacterial suspension and spread onto filters placed on IM agar containing 200μM AS. After 48h of incubation at 28°C, the filters were transferred to MM-hyg agar selection medium containing 300μg/mL cefotaxime (Wako Chemicals) to inhibit the growth of A. tumefaciens cell. The sample was frozen in liquid nitrogen and lyophilized. DNA was extracted based on the cetyl trimethyl ammonium bromide (CTAB) mini extraction method with slight modifications. The frozen samples were ground in a frozen pestle with liquid nitrogen. The finely ground mycelia were transferred into sterile centrifuge tubes and pre-warmed at 65°C. DNA extraction buffer [100mM Tris HCl (pH 8.0), 1.4M NaCl, 50mM EDTA (pH 8.0), and 2% CTAB] (Wako Chemicals USA, Inc.) was added to the mixture, mixed well and incubated with gentle shaking for 1h in a 65°C water bath. After incubation, an equal volume of chloroform: isoamyl alcohol (24:1, v/v) (Wako Chemicals USA, Inc.) was added gently to denature proteins and centrifuged for 10min at 15,670×g and 4°C. The aqueous phase was transferred into a sterile tube and DNA was precipitated for 20min with 0.6 volume of cold isopropanol (Wako Chemicals USA, Inc.) at 16,100×g and 4°C. The supernatant was poured off and the pellet was washed twice with cold 70% ethanol (Wako Chemicals USA, Inc.). Then, the pellet was air-dried at room temperature and suspended in TE buffer [10mM Tris HCl (pH 8.0), 1mM EDTA (pH 8.0)]. The sample was treated with RNase A (20mg/mL) (TaKaRa, Osaka, Japan) at 40ng/mL DNA and incubated for 1h at 37°C. Then, 1μL of genomic DNA from the knockout candidate was used as the template for PCR amplification using specific primers (ΔpksT-2 check F and ΔpksT-2 check R (Online Resource 1) and the Taq enzyme (TaKaRa, Osaka, Japan).
Single correct homologous integration through replacement of the resident ΔpksT-2 with ΔpksT-2:: hyg was confirmed via Southern blot analysis. The primers for the ΔpksT-2-S probe in the Online Resource 1 and its sequence are provided in the Online Resource 2.
Results and discussionIsolation of PKS genes via gene walking using degenerate primersThe PCR of the T. harzianum 88 genomic DNA using degenerate primers based on the KS- AT region yielded one fragment. The KA-series primers are described in Online Resource 1. The 700bp pksT-a-1 fragment was cloned into the pMD18-T Easy vector (Promega, Madison, WI, USA). The sequence was homologous to Penicillium marneffei PKSs (58% identity, GenBank accession no. XP_002151003) based on BLASTx searches against the NCBI/GenBank database. The pksT-a-1 fragment, amplified using the KAF1/KAR1 primers, was a putative non-reducing PKS fragment. However, PCR using other KA primer pairs for HP-reducing PKSs and PR-reducing PKSs did not produce any DNA fragment. We designed xKAF1 and xKAF2 as forward primers from the conserved regions of KS domains to obtain highly reducing PKSs. PCR using the xKAF1/KAR2 primers revealed one product with the expected size (800bp), pksT-b-1, which was highly similar to HP-reducing PKSs genes from Talaromyces stipitatus (57% similarity, GenBank accession no. XP_002478229.1) under BLASTx analysis. xKAF1 primer was derived from the aa sequence EM(S)HGTGTQ in the KS domain of HR-type PKSs, whereas xKAF2 primer was derived from EAEQMDPQ, which is conserved among PR PKSs, but amplification using xKAF2/KAR1 and KAR2 primers did not produce any viable DNA fragment. Baker et al.28 used a phylogenomic approach to study the PKS repertoire encoded in the genomes of Trichoderma reesei, T. atroviride and T. virens. They found only one PR PKS gene in T. atroviride. Therefore, T. harzianum may not express this class of PKS.
Aside from the two aforementioned sequences, no other sequences were obtained through gene walking even though different approaches were used to isolate further sequences. Therefore, a set of sense primers for 5′ end and 3′ end amplification was designed for gene walking (Online Resource 1). The sequences were obtained by overlapping the fragments isolated through degenerate PCR, and other fragments were generated using the gene walking primer sets. Finally, most of the 5′ and 3′ end regions of the putative PKSs were determined via gene walking amplification, and the putative β-ketoacyl synthase genes were identified through Blast searches. Two final PKS sequences, 7187bp (pksT-a) and 10,976bp (pksT-b), were obtained.
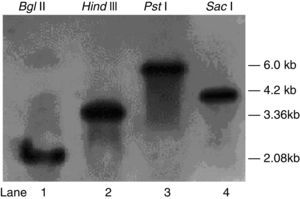
Detection of PKS gene copy numbersT. harzianum 88 genomic DNA was isolated using DNeasy Plant Maxi columns. Southern blot analysis with the DIG-labelled pksT-a probe revealed a single 2.08kb band after Bgl II cleavage and a 3.36kb band after Hind III cleavage (Fig. 1). Similarly, Southern blot analysis using the DIG-labelled pksT-b probe yielded a 6.0kb band after PstI cleavage and a 4.2kb band after SacI cleavage (Fig. 1). These findings demonstrate that pksT-a and pksT-b were present as single copies in T. harzianum 88. The iterative type of PKS (iPKS) only poses a single copy of each catalytic domain, however these can be deployed repeatedly during synthesis of a single polyketide molecule. And single copies of the genome are most structural genes that carry out a variety various functions. Therefore, identification of specific gene copy number is important for in-depth understanding of its biological function.
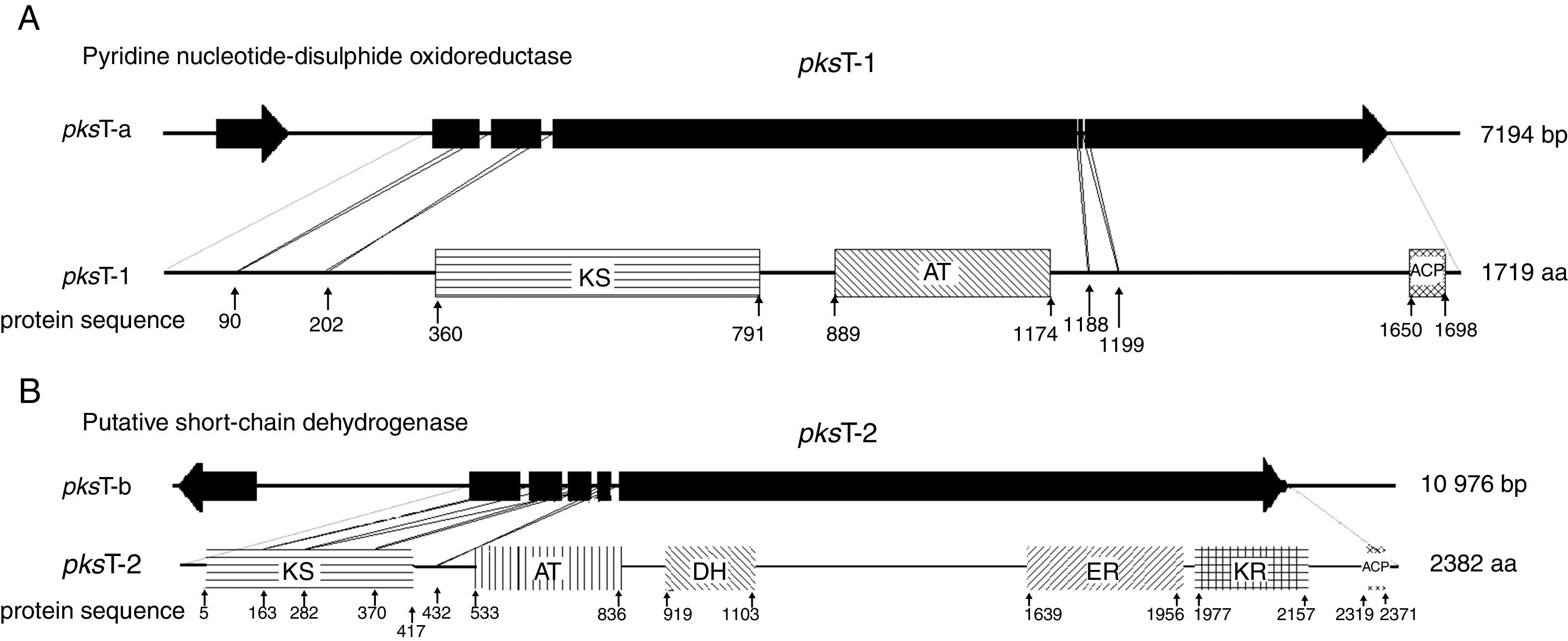
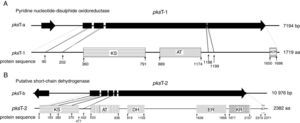
Domain organization of PKS genesAdditional domain analysis was performed on pksT-a and pksT-b sequences after using degenerate primers and gene walking to obtain the two PKS gene fragments, which included two complete PKS fragments (pksT-1 and pksT-2, respectively). A putative pyridine nucleotide–disulphide oxidoreductase gene clustered with pksT-1 on the sequenced pksT-a fragment (Fig. 2A). Another gene was clustered with pksT-2 on the pksT-b sequence, which was putatively identified as a short-chain dehydrogenase (Fig. 2B). The pksT-1 sequence was deposited in the NCBI GenBank database under the accession number JN556039.1.
Structure and domain organization of T. harzianum 88 gene clusters. (A) pksT-1 gene cluster structure in T. harzianum 88: two ORFs encoding a putative pyridine nucleotide-disulphide oxidoreductase, and pksT-1, respectively; pksT-1 gene structure: the four introns are represented by lines and exons are represented by blocks. Schematic organization of the domains: KS, β-ketoacyl synthase (360–791), AT, acyltransferase (889–1174), ACP, acylcarrier protein (1650–1698). (B) pksT-2 gene cluster structure in T. harzianum 88: two ORFs encoding a putative short-chain dehydrogenase, and pksT-2, respectively; pksT-2 gene structure: the four introns are represented by lines and the exons are represented by blocks. Schematic organization of the domains: KS (5–417), AT (533–836), DH (919–1103), ER (1639–1956), KR (1977–2157) and ACP (2319–2371). The approximate domain boundaries were determined using the iterative PKS domain search program (http://www.nii.ac.in/∼pksdb/sbsp ks/master.html).
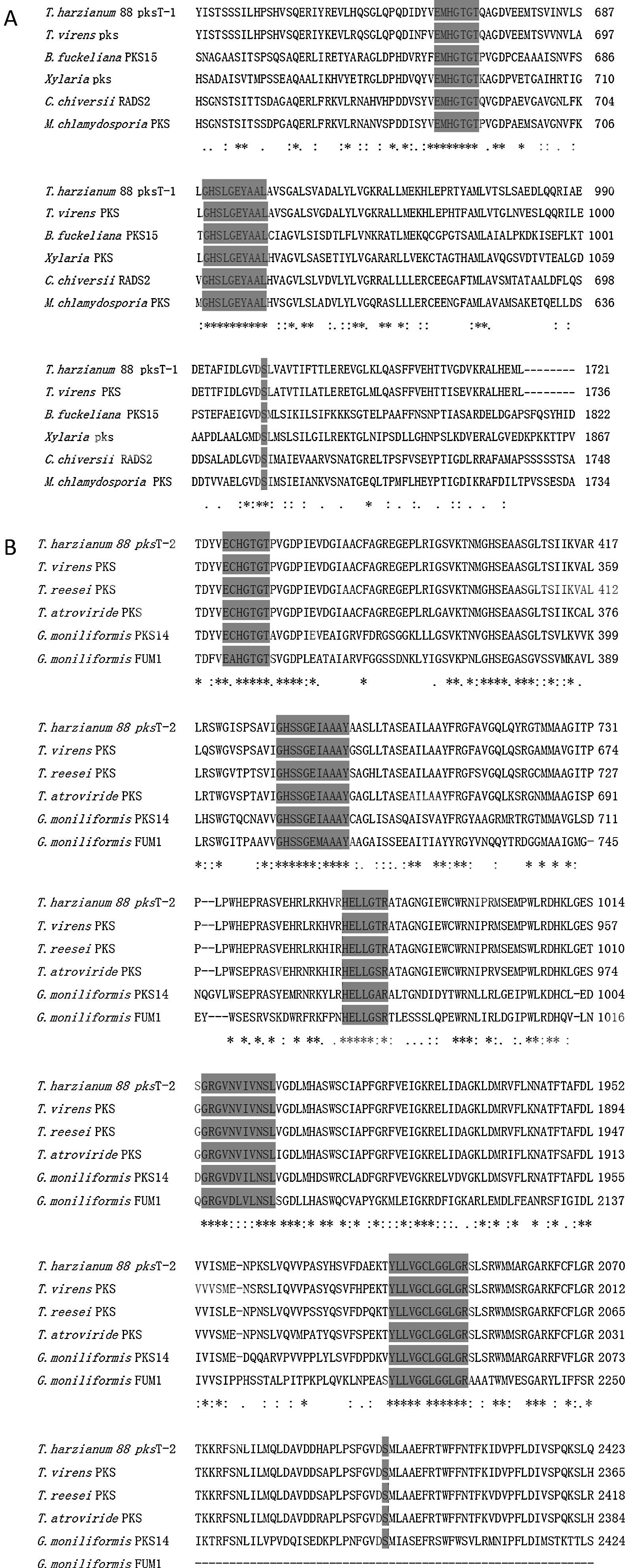
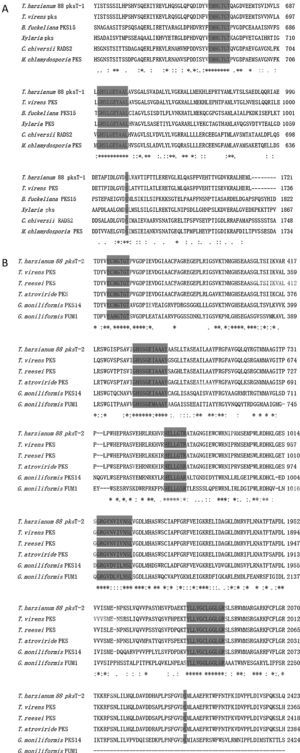
pksT-1, which contains 5669bp and encodes a 1719 aa protein with three exons (positions 31–299, 354–639, 699–5306 from the transcript inicial point), was confirmed via RT-PCR. The protein sequence of pksT-1 contained the entire ancestral domain structure of type I non-reducing PKSs based on the presence of conserved motifs: KS (β-ketoacyl synthase)/AT/ACP. Comparison between the pksT-1 protein sequence and the corresponding domains of other fungal PKSs showed conservation of the active site residues in KS (EMHGTGT), AT (GHSLGEYAAL) and ACP (GVDSLV) (Fig. 3A). The pksT-1 protein sequence was predicted to produce non-reduced non-cyclic PKS.20
Alignment of T. harzianum 88 predicted PKS active regions with closely related fungal PKSs. (A) Alignment of putative catalytic motifs found in the deduced amino acid sequence of T. harzianum 88 pksT-1 with those of T. virens PKS (EHK16308.1); B. fuckeliana PKS15 (BAE80697.1); Xylaria sp. BCC 1067 PKS (ABB90283.1); C. chiversii RADS2 (ACM42403.1); M. chlamydosporia (ACD39770.1). (B) Comparison with the highly conserved motifs of T. harzianum 88 pksT-2, T. virens (EHK18438.1), T. reesei QM6a PKS (EGR47100), T. atroviride (EHK47038.1), G. moniliformis PKS14 (AAR92221.1) and G. moniliformis FUM1 (AAR92218.1). Amino acid residues conserved among all sequences are marked with an asterisk. Positions with conserved substitutions are marked with two dots, and those with semi-conserved substitutions are marked with one dot. Highlighted regions indicate the catalytic amino acid residues in the active site.
Another PKS gene, pksT-2 is a 7901bp gene that encodes a 2382 aa protein. The presence of five exons (positions 228–718, 766–1074, 1127–1337, 1399–1522 and 1590–7603 from the transcript inicial point) was confirmed via RT-PCR. Six catalytic domains were identified in the pksT-2 protein sequence based on the presence of conserved motifs. These domains, in order from the N-terminus to the C-terminus were KS, AT, DH, ER, KR and ACP. Comparison of the pksT-2 protein sequence with the corresponding domains of other fungal PKSs showed conservation of the active site residues in KS (ECHGTGT), AT (GHSSGEIAAAY), DH (HELLGTR), ER (GRGVNVIVNSL), KR (YLLVGCLGGLGR), and ACP (FGVDSML) (Fig. 3B). The pksT-2 sequence was deposited in the NCBI GenBank database under accession number JN556040.1.
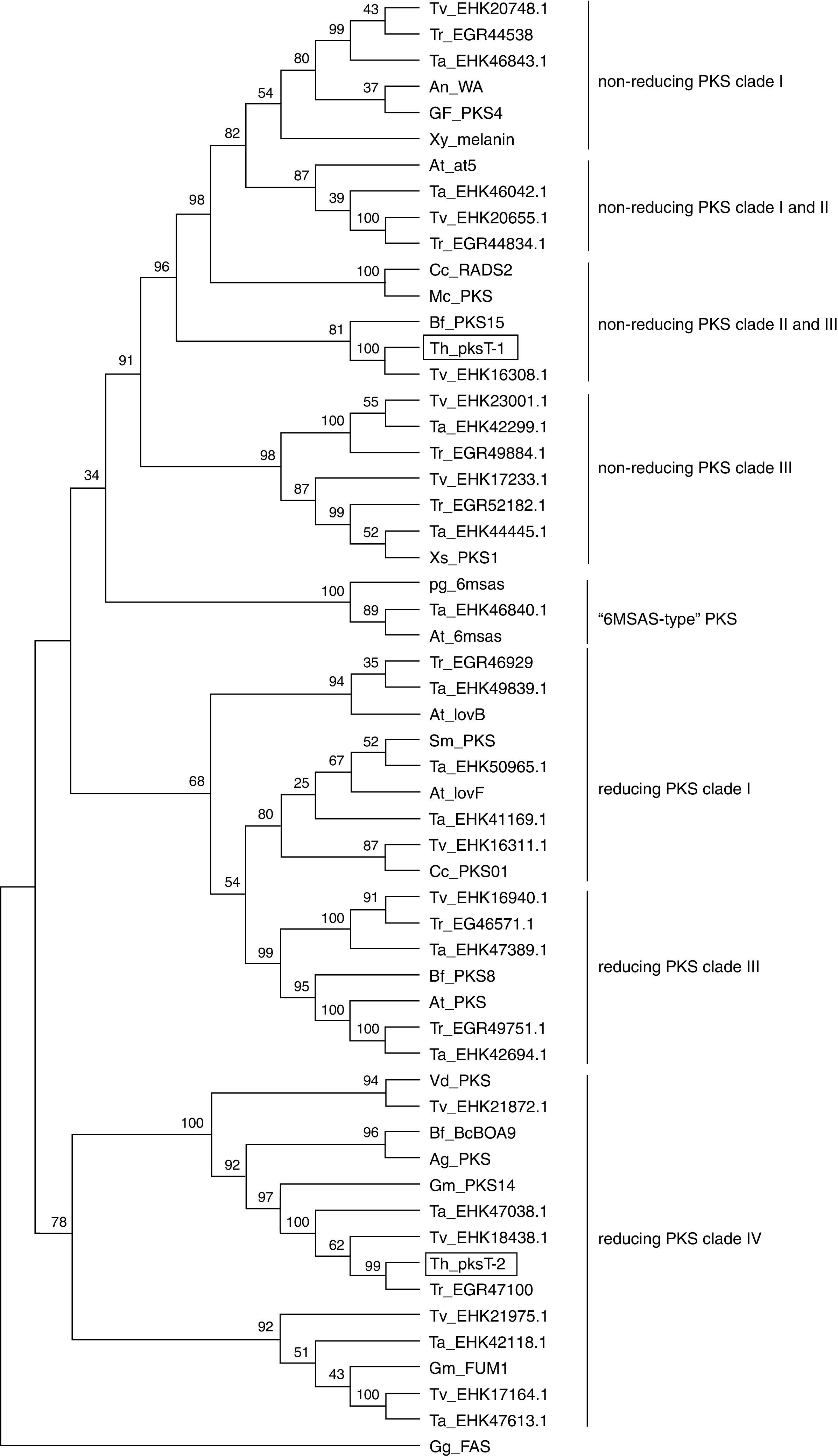
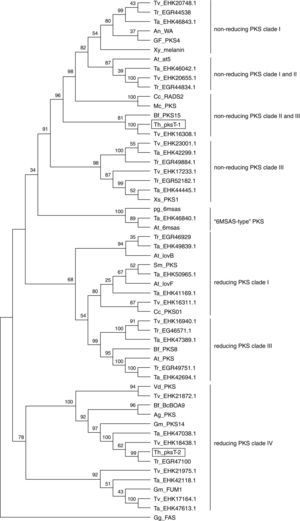
Phylogenetic analysis of the PKS fragmentsThe KS domain genealogy was used to infer the genealogy of type I fungal PKSs. Reliable sequences alignment of the proteins was impossible because of the insufficient similarity between members. The domains were group into previously established chemistry-based clades that represented non-reducing, partially reducing and highly reducing fungal PKSs based on a maximum parsimony phylogenetic analysis of the translated sequences. These three major clades were clustered into a larger clade of type I fungal PKSs, which is a sister clade of the FASs found in animals. In this present study, we chose several protein sequences from 3 Trichoderma spp., 15 filamentous fungal species and G. gallus to construct a phylogenetic tree.
The full aa sequences of the KS domains from several reducing/non-reducing fungal PKSs were employed to generate a phylogenetic tree (Fig. 4). Phylogenetic analysis revealed that pksT-1 was most closely related to the putative PKS from T. virens (80% similarity; GenBank accession no. EHK16308.1) and Botryotinia fuckeliana PKS15 (38% similarity; accession number AAR90251.1). pksT-1 generated a dependent subclade that was close to the fungal non-reducing PKS clade II and III with strong bootstrap values (100%). These homologous proteins also contain the same domain organization as KS-AT-ACP in type I fungal PKSs. The KS-AT-ACP domain structure is universal in fungi, such as in C. chiversii RADS2 and Metacordyceps chlamydosporia PKSs, which are involved in RAL biosynthesis. The phylogenetic analysis demonstrated that pksT-1 was more closely related to the PKSs involved in RAL biosynthesis than other known PKSs. RAL biosynthesis generally requires a non-reducing RADS2, a reducing PKS01, a halogenase and other cluster genes, but their functions are unknown.29 Alignment of the two protein sequences using the NCBI-Blast2p algorithm showed that one gene from T. virens (accession number EHK16311.1), which encodes a reducing PKS, is most closely related to C. chiversii PKS01. The other protein from T. virens (accession number EHK16308.1) is highly similar to C. chiversii RADS2 based on Blast2p alignment. Therefore, T. virens likely has a similar gene cluster structure for RAL biosynthesis.
Relationship of T. harzianum 88 PKSs to fungal type I iterative PKSs. The tree was constructed with type I iterative PKS protein sequences. β-Ketoacyl synthase of G. gallus FAS was used as the out-group. A multiple sequence alignment was performed using the CLUSTALW (1.83) program, and the tree was generated with the MEGA (4.0) program. The indicated scale represents 0.1 amino acid substitution persite. PKSs of T. harzianum 88 are indicated in bold and framed with rectangles. Abbreviations, genus and species names and the NCBI ENTREZ accession numbers of the 55 fungal PKS sequences and the FAS sequence from G. gallus are given in abbreviation table.
pksT-2 generated another subclade with very strong bootstrap values (99%; Fig. 4) close to the fungal reducing PKS clade IV. These homologous clones also contain the same domain organization as KS-AT-DH-ER-KR-ACP in the type I PKS genes. Phylogenetic analysis using the amino acid sequences of the KS domain revealed that pksT-2 is most closely related to the putative PKS from T. virens (86% sequence identity; accession number EHK18438.1) and G. moniliformis (33% sequence identity; accession number AAR92218.1). The KS domain analysis showed that pksT-2 may be involved in fumonisin production based on an analysis of the NaPDoS database. Reducing PKS subclade IV, and subclades I and II may or may not have a conserved methyltransferase domain. The only characterized PKS in this subclade was G. moniliformis FUM1, which contains the KS-AT-DH-(C-MT)-ER-KR-ACP domains and forms the linear PK precursor of the toxin fumonisin.30 The FUM1 expression levels was highly correlated with fumonisins based on real-time RT-PCR, which suggests that real-time RT-PCR is suitable for studying the influence of abiotic and biotic factors on the expression of these genes.31,32 Cox et al.33 found that the C-MeT domain transfers a methyl group after the first and second rounds of iteration, but it is inactive in the final round along with the ER domain. However, many of the predicted PKSs in the PKS clade producing reduced PKs have high divergence and presumably nonfunctional MT domains.21pksT-2 does not contain the C-MT domain compared with domains of G. moniliformis FUM1, which may synthesize a compound similar to the linear PK precursor of fumonisin.
The NCBI ENTREZ accession numbers of the 55 fungal PKS sequences and a FAS sequence from G. gallus are shown in abbreviations table.
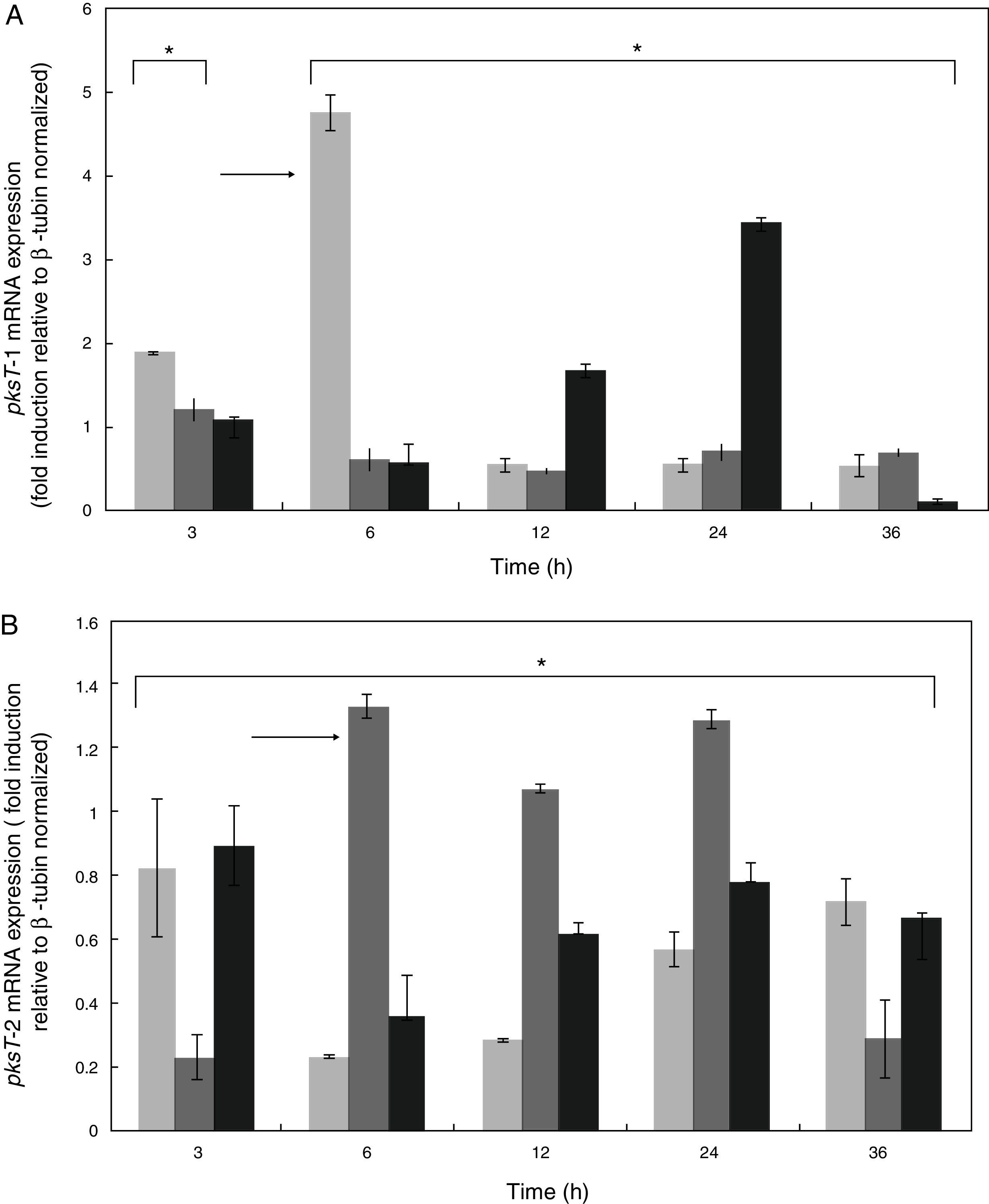
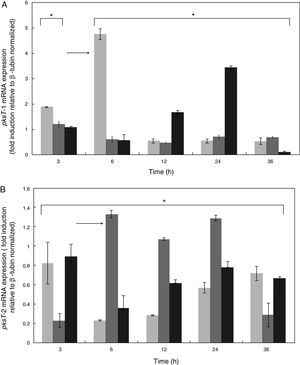
Expression patterns of pksT-1 and pksT-2 genesWe studied the effect of T. harzianum 88 confrontation with different fungal pathogens (S. sclerotiorum, R. solani and F. oxysporum) in the mRNA levels of pksT-1 and pksT-2 to elucidate the pksT genes expression. In the present study, fungal pathogens induction of two genes was noticeably high in T. harzianum 88 compared with the untreated controls.
pksT-1 expression was highly upregulated by S. sclerotiorum, increasing 1.88-fold within 3h and peaking at 4.74-fold within 6h. The pksT-1 expression was decreased at 12, 24 and 36h after induction. Significant changes in pksT-1 mRNA levels were observed when T. harzianum 88 was challenged with S. sclerotiorum (p≤0.01). R. solani challenge induced the highest pksT-1 mRNA levels at 1.22-fold within 3h. However, the pksT-1 mRNA levels decrease after 6, 12, 24 and 36h, and the lowest level was observed at 12h. These significant changes in pksT-1 mRNA levels were also observed when T. harzianum 88 was challenged with R. solani (p≤0.01). pksT-1 was also upregulated 1.07-fold by the F. oxysporum challenge compared with the untreated control at 3h, but the increase was not statistically significant (p>0.05). The increases in pksT-1 expression at 6h and at 12h were significant, reaching its peak at 3.42-fold after 24h, and significantly decreasing after 36h (p≤0.05; Fig. 5A).
Time course expression analyses of PKS genes responding to confrontation with R. solani, S. sclerotiorum or F. oxysporum. X-axis: time points of confrontation with phytopathogens; Y-axis: mRNA expression (fold induction relative to β-tubin normalized). The fold change in PKS gene expression was calculated using the formula 2−ΔΔCT. White bars: expression responding to S. selerotiorum; horizontal bars: expression responding to R. solani; Black bars: expression responding to F. oxysporum. *Significant difference at 0.05 level (p<0.05) by comparing T. harzianum 88 which was confrontation with three fungal plant pathogens and non- confrontation with three fungal plant pathogens. Arrows indicate the highest expression of PKS genes responding to different phytopathogens.
The S. sclerotiorum-induced pksT-2 expression increased to almost 0.8-fold compared with the untreated samples at 3h. The pksT-2 expression decreased after 6h, but increase from 12h to 36h (p<0.05; Fig. 5B). We also observed the highest increase in pksT-2 expression (1.33-fold) at 6h after R. solani challenge (p<0.05; Fig. 5B). Similarly, F. oxysporum increased pksT-2 expression by ∼0.93-fold compared with the untreated samples (p<0.05 at 3, 6, 12, 24 and 36h; Fig. 5B). The RT-qPCR analysis showed that the fungal pathogens induce pksT-2 gene expression. In summary, fungal pathogens induce the transcription of pksT genes in T. harzianum 88.
These results were similar to that of numerous polyketide synthases, which are upregulated in creeping bentgrass infected with Sclerotinia homoeocarpa.34 By contrast, the PcPKS2 transcript levels in the leaves are highly upregulated by pathogens.35 6msas activates disease resistance pathways by enhancing the accumulation of PR proteins and resistance to tobacco mosaic virus.36 PKS/non-ribosomal peptide synthetase (NRPS) hybrid enzyme is also involved in Trichoderma–plant interactions that induce defence responses.37 In addition, T. harzianum is a mycoparasitic filamentous fungus that produces several antifungal compounds when challenged with fungal pathogens to contribute to fungistasis. These antifungal compounds may participate in the biocontrol mechanism of Trichoderma.
In addition, PKS expression in fungi has not been studied in relation to pathogenesis and Trichoderma. The results indicate that with at least one of the three fungal pathogens causes the differential regulation of the two PKS genes. However, these genes generally display different expression patterns when Trichoderma encounters different fungal pathogens, which suggests that T. harzianum generates specific response patterns to different challenges.
Construction of the ΔpksT-2 disruptant strainThe effect of the hygromycin B dose on the regeneration frequency of conidia was determined before transformation. Hygromycin B at 100μg/mL completely inhibited the regeneration of untransformed protoplasts. Therefore, growth at this concentration was used as the selection criterion for hygromycin B-tolerant transformants (Fig. 6).
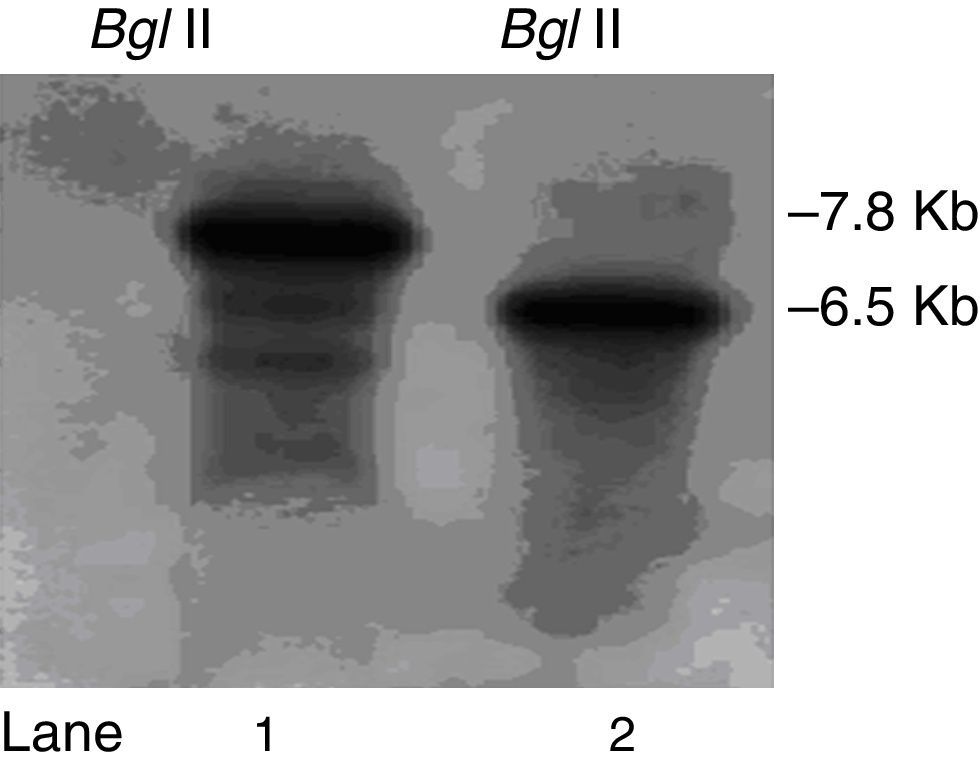
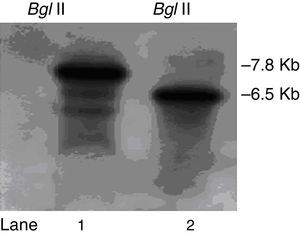
We constructed a ΔpksT-2 disruptant strain to examine the function of pksT-2 in T. harzianum. A. tumefaciens-mediated transformation obtained six candidate colonies. Subsequently, these colonies were screened via colony PCR using the primers ΔpksT-2 check R and ΔpksT-2 check F (Online Resource 1). We further confirmed the ΔpksT-2 disruptant strain through Southern blot analysis. These analyses revealed the expected hybridization signal at 7.8kb and at 6.5kb in the digested genomic DNA isolated from the wild-type T. harzianum 88 and the ΔpksT-2 candidate transformant with BglII (Fig. 6). These results indicate successful gene replacement at the resident ΔpksT-2 locus.
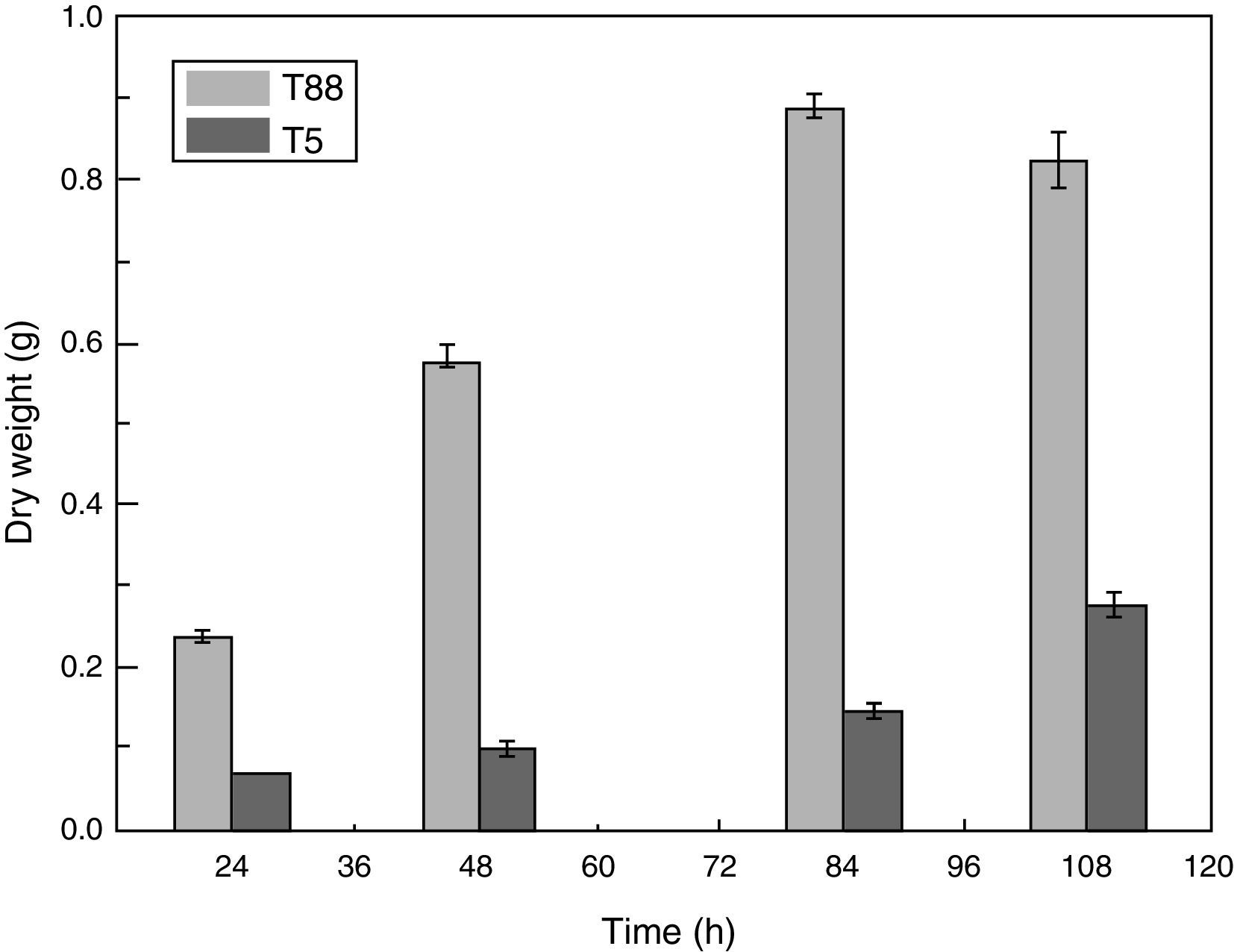
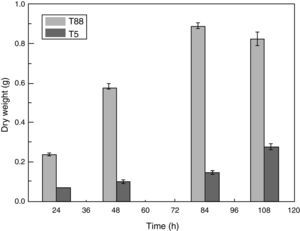
Characterization of the pksT-2 disruptant strainWe examined the pksT-2 disruptant strain to determine its growth characteristics. Cell dry weight of wild-type and pksT-2 disruptant strains increased with incubation time, and which of wild type T. harzianum 88 reached the highest weight at 84h. However, based on the data obtained from the cell growth curves of T. harzianum 88 strain in our lab (data not shown), its cell has entered a decline phase at 108h, At this point the cell dry weight is slightly lower at 108h than at 84h. Furthermore, cell dry weight of pksT-2 disruptant strain growth slower than wild type T. harzianum 88, and which reached the highest weight at 108h, but only 33% of the dry weight of the wild-type strain cell. The result showed that the growth rate of the strain harbouring the pksT-2 disruptant on solid and liquid media was significantly lower than that of the wild-type strain (Fig. 7).
Growth rates of the wild-type strain and pksT-2 disruptant. The wild-type strain and pksT-2 disruptant were inoculated in liquid MM medium (1×106conidia/mL) and grown at 28°C for 24, 48h and 72h, 150rpm/min respectively. The data represent the mean±S.D (n=3). Black bar: wild-type; white bar: pksT-2 disruptant.
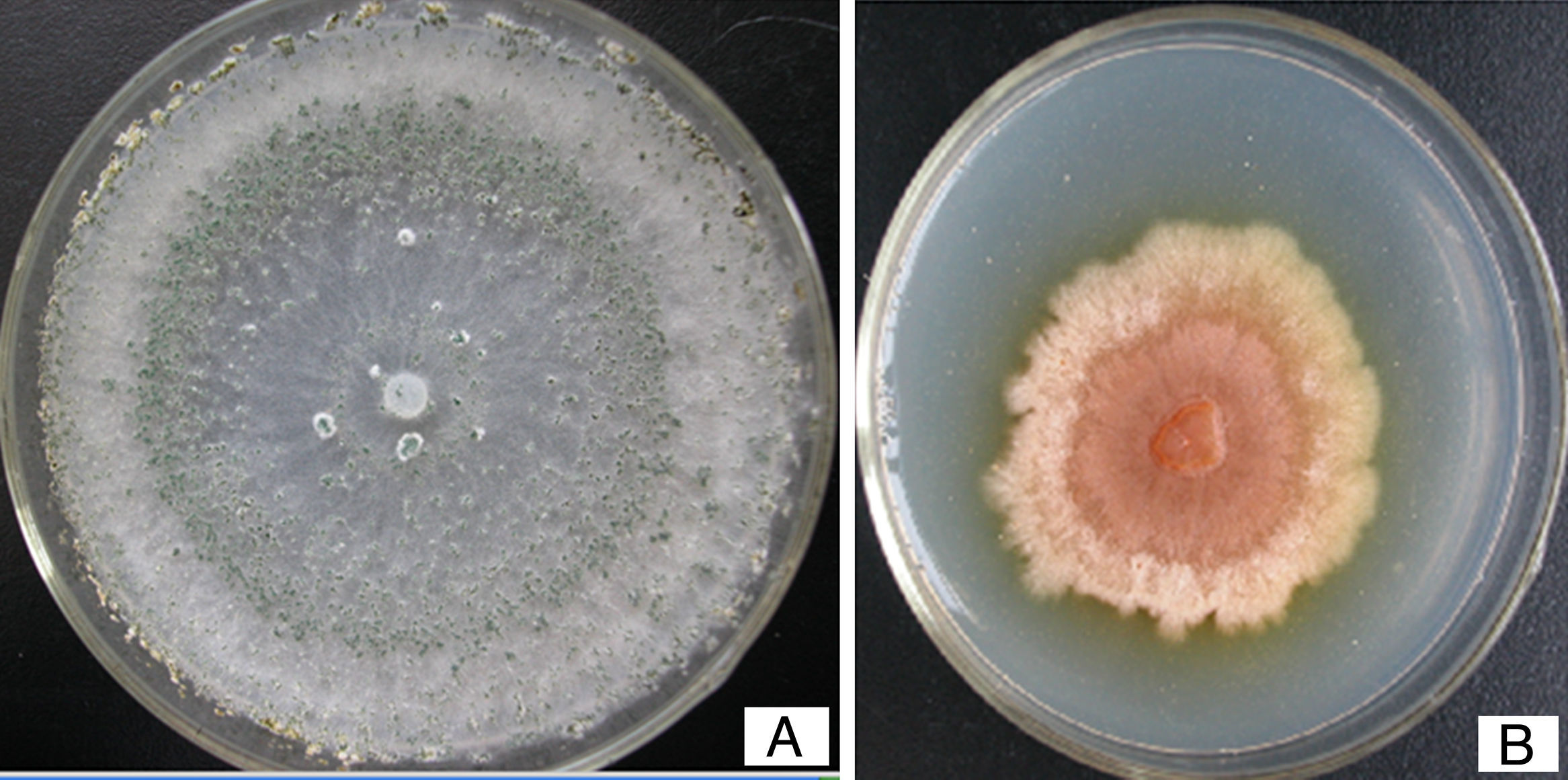
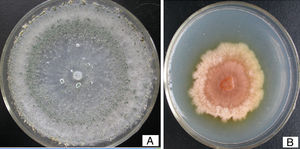
We observed the phenotype of the pksT-2 disruptant strain to confirm the function of pksT-2 and to evaluate the gene targeting efficiency. T. harzianum 88 produced green conidia (Fig. 8A). Under the same growth conditions T. harzianum 88 harbouring the pksT-2 disruptant strain displayed a pink conidial phenotype (Fig. 8 B), and the yellow conidiation spread into solid medium. This result indicates that inactivation of the pksT-2 gene influences the conidial pigmentation of T. harzianum 88. Polyketides are involved in the biosynthesis of important natural products, such as fungal pigments,38 antibiotics, and a variety of therapeutic compounds.39 The function of a number of genes in T. harzianum 88 genomic DNA that are involved in secondary metabolic pathways, such as PKS and NRPS are unknown. Thus, T. harzianum 88 with the pksT-2 disruptant will be useful for gene disruption experiments to analyse gene function.
ConclusionsIn conclusion, pksT-1 and pksT-2 were isolated from T. harzianum 88. Although secondary metabolites are unrelated to pksT-1 and pksT-2, analysis of their domain organization implied that pksT-1 provides a side chain in RAL biosynthesis and pksT-2 is involved in compound biosynthesis similar to fumonisin biosynthesis. The expression levels of the pksT-1 and pksT-2 genes were evaluated via real-time RT-PCR during challenge with different fungal pathogens (S. sclerotiorum, R. solani and F. oxysporum). The results indicated that pksT-1 and pksT-2 are differentially regulated when T. harzianum 88 is challenged with at least one of three fungal pathogens. A pksT-2 disruptant strain was obtained through gene functional inactivation, and its growth rate was slower than that of the wild type. The pksT-2 disruptant strain showed that the pksT-2 gene encodes a key enzyme which may be involved with pigmentation.
Future studies will focus on the heterologous expression of pksT-1 and pksT-2 to confirm their functions. These studies will provide a basis for the sustainable application of T. harzianum 88 secondary metabolites in pharmaceutical treatment and biocontrol of plant pathogens.
Conflicts of interestThe authors declare no conflicts of interest.
Project supported by the National Science Foundation for Distinguished Young Scholars of China (Grant No. 31501839).