The microorganism–microorganism or microorganism–host interactions are the key strategy to colonize and establish in a variety of different environments. These interactions involve all ecological aspects, including physiochemical changes, metabolite exchange, metabolite conversion, signaling, chemotaxis and genetic exchange resulting in genotype selection. In addition, the establishment in the environment depends on the species diversity, since high functional redundancy in the microbial community increases the competitive ability of the community, decreasing the possibility of an invader to establish in this environment. Therefore, these associations are the result of a co-evolution process that leads to the adaptation and specialization, allowing the occupation of different niches, by reducing biotic and abiotic stress or exchanging growth factors and signaling. Microbial interactions occur by the transference of molecular and genetic information, and many mechanisms can be involved in this exchange, such as secondary metabolites, siderophores, quorum sensing system, biofilm formation, and cellular transduction signaling, among others. The ultimate unit of interaction is the gene expression of each organism in response to an environmental (biotic or abiotic) stimulus, which is responsible for the production of molecules involved in these interactions. Therefore, in the present review, we focused on some molecular mechanisms involved in the microbial interaction, not only in microbial–host interaction, which has been exploited by other reviews, but also in the molecular strategy used by different microorganisms in the environment that can modulate the establishment and structuration of the microbial community.
Microbial interactions are crucial for a successful establishment and maintenance of a microbial population. These interactions occur by the environmental recognition followed by transference of molecular and genetic information that include many mechanisms and classes of molecules. These mechanisms allow microorganisms to establish in a community, which depending on the multi-trophic interaction could result in high diversity. The result of this multiple interaction is frequently related to pathogenic or beneficial effect to a host. In humans, for example, the microbial community plays an important role in protection against diseases, caused by microbial pathogens or physiological disturbances. Soils microbial communities also play a major role in protecting plants from diseases and abiotic stresses1 or increasing nutrient uptake.
Microorganisms are rarely encountered as single species populations in the environment, since studies in different habitats has shown that an enormous richness and abundance variation are usually detected in a small sample, suggesting that microbial interactions are inherent to the establishment of populations in the environment, which includes soil, sediment, animal, and plants, including also fungi and protozoa cells. The many years of coevolution of the different species lead to adaptation and specialization and resulted in a large variety of relationships that can facilitate cohabitation, such as mutualistic and endosymbiotic relationships, or competitive, antagonistic, pathogenic, and parasitic relationships.2
Many secondary metabolites have been reported to be involved in the microbial interactions. These compounds are usually bioactive and can perform important functions in ecological interactions. A widely studied mechanism of microbial interaction is quorum sensing, which consists in a stimuli-response system related to cellular concentration. The production of signaling molecules (auto-inducers) allows cells to communicate and respond to the environment in a coordinated way.3 During interaction with the host cells, microbial-associated molecular patterns (PAMP or MAMP – microbial-associated molecular pattern) are conserved throughout different microbial taxon allowing to increase the fitness during interaction with plant or animal cells4 and regulating the microbial interactions with different hosts (Table 1).
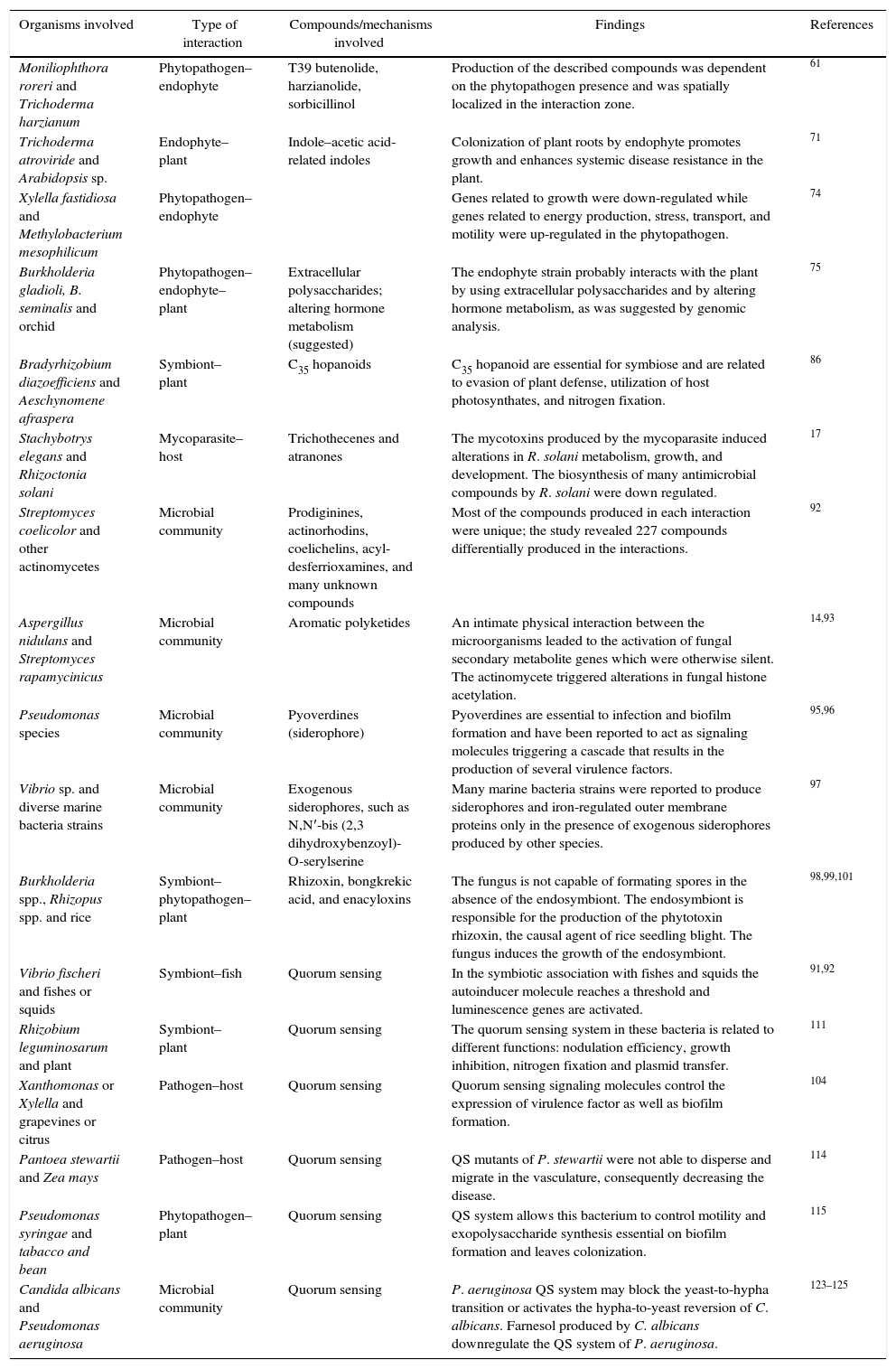
Microbial interaction studies.
| Organisms involved | Type of interaction | Compounds/mechanisms involved | Findings | References |
|---|---|---|---|---|
| Moniliophthora roreri and Trichoderma harzianum | Phytopathogen–endophyte | T39 butenolide, harzianolide, sorbicillinol | Production of the described compounds was dependent on the phytopathogen presence and was spatially localized in the interaction zone. | 61 |
| Trichoderma atroviride and Arabidopsis sp. | Endophyte–plant | Indole–acetic acid-related indoles | Colonization of plant roots by endophyte promotes growth and enhances systemic disease resistance in the plant. | 71 |
| Xylella fastidiosa and Methylobacterium mesophilicum | Phytopathogen–endophyte | Genes related to growth were down-regulated while genes related to energy production, stress, transport, and motility were up-regulated in the phytopathogen. | 74 | |
| Burkholderia gladioli, B. seminalis and orchid | Phytopathogen–endophyte–plant | Extracellular polysaccharides; altering hormone metabolism (suggested) | The endophyte strain probably interacts with the plant by using extracellular polysaccharides and by altering hormone metabolism, as was suggested by genomic analysis. | 75 |
| Bradyrhizobium diazoefficiens and Aeschynomene afraspera | Symbiont–plant | C35 hopanoids | C35 hopanoid are essential for symbiose and are related to evasion of plant defense, utilization of host photosynthates, and nitrogen fixation. | 86 |
| Stachybotrys elegans and Rhizoctonia solani | Mycoparasite–host | Trichothecenes and atranones | The mycotoxins produced by the mycoparasite induced alterations in R. solani metabolism, growth, and development. The biosynthesis of many antimicrobial compounds by R. solani were down regulated. | 17 |
| Streptomyces coelicolor and other actinomycetes | Microbial community | Prodiginines, actinorhodins, coelichelins, acyl-desferrioxamines, and many unknown compounds | Most of the compounds produced in each interaction were unique; the study revealed 227 compounds differentially produced in the interactions. | 92 |
| Aspergillus nidulans and Streptomyces rapamycinicus | Microbial community | Aromatic polyketides | An intimate physical interaction between the microorganisms leaded to the activation of fungal secondary metabolite genes which were otherwise silent. The actinomycete triggered alterations in fungal histone acetylation. | 14,93 |
| Pseudomonas species | Microbial community | Pyoverdines (siderophore) | Pyoverdines are essential to infection and biofilm formation and have been reported to act as signaling molecules triggering a cascade that results in the production of several virulence factors. | 95,96 |
| Vibrio sp. and diverse marine bacteria strains | Microbial community | Exogenous siderophores, such as N,N′-bis (2,3 dihydroxybenzoyl)-O-serylserine | Many marine bacteria strains were reported to produce siderophores and iron-regulated outer membrane proteins only in the presence of exogenous siderophores produced by other species. | 97 |
| Burkholderia spp., Rhizopus spp. and rice | Symbiont–phytopathogen–plant | Rhizoxin, bongkrekic acid, and enacyloxins | The fungus is not capable of formating spores in the absence of the endosymbiont. The endosymbiont is responsible for the production of the phytotoxin rhizoxin, the causal agent of rice seedling blight. The fungus induces the growth of the endosymbiont. | 98,99,101 |
| Vibrio fischeri and fishes or squids | Symbiont–fish | Quorum sensing | In the symbiotic association with fishes and squids the autoinducer molecule reaches a threshold and luminescence genes are activated. | 91,92 |
| Rhizobium leguminosarum and plant | Symbiont–plant | Quorum sensing | The quorum sensing system in these bacteria is related to different functions: nodulation efficiency, growth inhibition, nitrogen fixation and plasmid transfer. | 111 |
| Xanthomonas or Xylella and grapevines or citrus | Pathogen–host | Quorum sensing | Quorum sensing signaling molecules control the expression of virulence factor as well as biofilm formation. | 104 |
| Pantoea stewartii and Zea mays | Pathogen–host | Quorum sensing | QS mutants of P. stewartii were not able to disperse and migrate in the vasculature, consequently decreasing the disease. | 114 |
| Pseudomonas syringae and tabacco and bean | Phytopathogen–plant | Quorum sensing | QS system allows this bacterium to control motility and exopolysaccharide synthesis essential on biofilm formation and leaves colonization. | 115 |
| Candida albicans and Pseudomonas aeruginosa | Microbial community | Quorum sensing | P. aeruginosa QS system may block the yeast-to-hypha transition or activates the hypha-to-yeast reversion of C. albicans. Farnesol produced by C. albicans downregulate the QS system of P. aeruginosa. | 123–125 |
Much attention has been given to researches on microbial interactions in the human health field. The microbial interactions are crucial for the successful establishment and maintenance of colonization and infection. Additionally, antimicrobial host defenses and environmental factors also play essential roles. Microorganism communication enables the population to collectively regulate the gene expression in response to host and environmental signals, produced by the same or even by different species. This results in a coordinate response in the microbial population, achieving successful pathogenic outcomes that would not be accomplished by individual cells.5–7
Consequently, knowledge on the mechanisms involved in the microbial interactions can be a key to developing specific agents that can avoid or disturb microorganism communication during infection and consequently act to decrease the defensive and offensive qualities of the pathogen. Thus, the study of these mechanisms can contribute to the understanding of the microbial pathogenesis and to the development of new antimicrobial drugs.5,8
In addition, microbial interactions occurring in human host can also be benefic and some diseases are often related to imbalances in the healthy microbiota. Therefore, studies on the healthy microbial community in the host are also relevant as it can lead to disease prediction and its appropriate therapies.9–11
Microbial interactions also deserve attention from the natural products discovery field. Secondary metabolite clusters that are silent under laboratory growing conditions, can be activated by simulating the natural habitat of the microorganism. It has been reported that co-cultivation with others microorganisms from the same ecosystem can induce the activation of otherwise silent biosynthetic pathways leading to the production and identification of new natural products.12–16 Furthermore, this knowledge can also be applied to genetic engineering of phytopathogens antagonists/parasites aiming to an enhanced biological control.17
In this review, we focused on the molecular mechanisms involved in many microbial interactions, involving intra and interspecies microbial interactions and the microorganism interaction with the host.
Organisms involvedMicroorganisms rarely occur as single species populations and are encountered in many hosts/environments, thus there is a large variety of types of microbial interactions concerning the organisms involved. Bacteria–bacteria, fungus–fungus, bacteria–fungus, fungus–plant/animal, bacteria–plant/animal and bacteria–fungus–plant/animal interactions, including parasitic, mutualistic interactions involve many mechanisms that have been described, allowing to develop strategies to manipulate these interactions, which could result in increased host fitness or new metabolite production. According to van Elsas et al.,18 the establishment of a new species (invader) in an environment depends on the characteristic of the local microbial community. In general, ecosystems that lost species diversity present less ability to resist to an invader, since present more available niche that could be occupied by indigenous species. In addition, during the niche occupation, the invader should interact with species present in this environment.
The mechanisms involved in archaeal interactions are largely unknown, although they are very important in the archaeal communities, production of methane in landfills,19 archaea in soil and rhizosphere ecosystems,20 thermophilic archaea in bioleaching process,21 for example. Virus interactions with its host are also very important since viruses are responsible for many diseases in a variety of hosts, and also, modulating the bacterial community by infecting dominant species. Host-virus communication is related to RNA-based mechanisms such as microRNAs.22,23 The microorganisms addressed in the present reviewed comprise fungi and bacteria, we did not focus on virus or archaea.
Fungi and bacteria interactions are widely studied, although the molecular mechanisms involved in the interactions are often not completely understood. They interact with a wide range of different organisms – plants, humans and other animals, among others – in different environments, as we describe in this present review, and present many biotechnological applications, such as in food processing, bioremediation, medicine, and biocontrol. In addition, fungal–bacterial association forms a physically and metabolically interdependent conglomerate that presents distinct properties which are biotechnology relevant, especially considering the natural product discovery and synthetic biology field.1,24
There are many microbe–host interactions, which can be related to beneficial or pathogenic interactions in plants and animals. In these interactions, the microbial cells may be found in biofilm or planktonic state, which result in different genetic and physiological states.
Plant-associated microorganisms (endophytic and rhizosphere environment) are able to promote plant growth by producing phytohormones, improving biofertilization, bioremediation, and reducing biotic (disease) and abiotic stress.25 Root-associated endophytes are able to produce phytohormones as auxins and gibberellins promoting plant growth. Considering biofertilization, rhizosphere bacteria are able to fix atmospheric nitrogen, produce siderophore for iron acquisition and mycorrhizal fungi is able to solubilize phosphorus making it available to plant host.26
The control of plant stress is exemplified by the production of ACC deaminase that is responsible for the decrease of ethylene levels by cleaving its precursor 1-aminocyclopropane-1-carboxylate (ACC) to ammonia and 2-oxobutanoate, lowering ethylene signaling and this way alleviating plant stress.27 A Burkholderia phytofirmans PsJN mutant in the ACC deaminase gene losses the ability to promote root elongation.28 Besides that, the inoculation of a mutualistic bacteria can also affect plant fitness by increasing photosynthetic rate, CO2 assimilation, and chlorophyll content.29,30
The presence genes related to plant growth promoting were addressed in studies comparing the genome of endophytes and pathogens, revealing that pathogens present genes involved in degradation and host invasion while mutualists present genes related to help in stress amelioration, encoding nitrogen fixation proteins and ribulose bisphosphate carboxylase/oxygenase (RubisCO) proteins.26
Microbiome interaction with its hostHumanThe human microbiome evolves from birth to elderly, resulting in microbial richness and diversity shifts over the whole life, modulating the immune system and physiological and morphological aspects of the host. Although some bacteria may be found in amniotic liquid with or without disease symptoms,31–33 the development of the human microbiome is studied from birth until this microbial community becomes adult-like. During the whole life, the human microbiome suffers imbalances that have been associated with several kinds of disease, such as asthma, obesity, diabetes, cancer and inflammatory problems in many body sites. The microbiome imbalance is referred as dysbiosis and may result in functional disease or may be caused by a disease or disease treatment. For a better comprehension of the association between intestinal microbial dysbiosis and pediatric diseases the Arrieta et al.,34 review present an important description of the microbial composition and shifts associated with the age.
For the development of this microbial community, the species that will compose this microbiota must show the ability to occupy the available niches and interact with the established microorganism and with host tissues. For this, microbes have to shape their environment by secretion products from their metabolism in a process called niche construction. During this process, the niche is constructed when the microorganisms manage the nutrient available and possible competitors by producing extracellular enzymes, antimicrobial compounds or activating or inhibiting the host immune system.35 Among this molecules, bacteriocin (peptides produced by a bacterium, that has an immunity mechanisms) active against other bacteria seems to be ubiquitous in bacteria and archaea domain and associated with niche construction, since these ribosomal peptides may work facilitating the introduction and/or dominance of a producer into an already occupied niche, or directly inhibiting competing strains or pathogens during gut colonization of working as signaling peptide (cross-talking) or signaling the host by interaction with receptors for immune system.36
Therefore, it is believed that the evolution of a microbial community in the host may be further related to an intrinsic characteristic of this community and the ability of the microbial species to construct their niche. The intrinsic aspects are associated to functional redundancy of the native community, reducing the available niches and the niche construction the ability of the invader to manage the environment (biotic and abiotic characteristics) in a social evolutionary behavior, resulting in a shaped environment that allows the establishment of the microbial colonizer into the host.
During establishment in the gut, microorganisms interact with the host cells expressing adhesive molecules on their surface, promoting interaction with cell receptors and triggering host responses. The most important adhesive structures are pili and fimbrial adhesins in Gram-negative bacteria, but others monomeric surface bound adhesive proteins has been largely identified.37 Although the regulation of adhesins has been studied mainly in pathogens, is believed that the same strategy has been used by commensal species. The chaperone-usher pathway has been an important system to assembly pilus adhesins of enteric pathogens, but others such as type IV pili, trimeric autotransporter adhesins (TAA) family, adhesive amyloids (Curli) (Gram-negative bacteria) and Sortase assembled Pili and putative head-stalk-type adhesin (Gram-positive bacteria) are secreted and assembled by Sec-dependent transporter.37 These adhesins allow the physical contact between the bacterial cells and host. This interaction mediated by both pilus-associated and non-pilus-associated adhesins with host receptors trigger host inflammatory responses. In addition, this attachment onto eukaryotic cells allows bacteria suppress the host defense by secretion of effector proteins into the host by secretion systems.37 Thus, the gut colonization begins rapidly after birth with the microorganism entry by ingestion and keeps going by shaping the environment and attachment onto the host cells or living into the gut lumen.
In addition, during the establishment into the host, gut microbiota may trigger tolerance or inflammatory response in the host. Some Lactobacillus spp. have the ability to induce rheumatoid arthritis by activating TLR (Toll-like receptor) 2 and TLR4 followed by increasing of TH1 and TH17 activity and decreasing TReg-cells function. The production of pro-inflammatory cytokines (IL-17) and endogen TLR4 agonist mediate joint inflammation by stimulating plasma cells to produce arthritogenic autoantibodies. However, some commensal bacteria, such as Bacteroides fragilis are able to activate pro-tolerogenic machinery, the PSA, a cell wall component, induce activation of TReg-cells and IL-10 production and repression of TH17-cell, avoiding uncontrolled inflammation.38 Cell wall components, such as peptidoglycans (PGN) may also spread into the host and be recognized by pattern recognition receptor (PRRs). The recognition of this PGN may trigger, not only a host immune response, but also host metabolism and behavior.39
During bacterial growth, PGN is degraded and although bacterial PGN recycling pathway tries to reduce the bioavailability of soluble fragments (preventing detection by the host)39 fragments (muropeptides) could disseminate systemically, activating receptors far from the gut. In fact, receptors (Nod1 and Nod2) that recognize these PGN fragments are broadly distributed into the human and animal bodies. In addition, in rats that present sleep deprivation, the bacterial translocation from the intestine to the mesenteric lymph nodes was observed.40 and in previous studies, it was observed that muramyl peptides may induce a somnogenic response after brain ventricule41 or intravenous or injection.42 These results suggest that sleep deprivation could induce bacterial translocation, which could be a source of muramyl peptides for sleeping induction.39 These results suggest that the host behavior could modulate the interaction with the microbial community, which in turn contribute to shifts in the host physiology.
Soil and plantAll organisms are inhabited by microorganisms including archaea, bacteria, fungi and viruses; this microbiota presents a key role in host health and development.43,44 The microbiome associated with plants is considered its second genome. It is determinants for plant health, growth, fitness and consequently productivity.45 Where each environment associated with the plant: rhizosphere, endosphere, and phyllosphere present a specific microbial community with specific functions.43
These culture-independent methods show that plant microbiome can reach densities greater than the number of plant cells and also greater expressed genes than the host cells. Metagenomics analysis using next-generation sequencing technologies shows that only 5% of bacteria have been cultured by current methods, revealing how many microorganisms and its functions remains unknown.25
The first step in plant–microbe interaction is microbial recognition of plant exudates in the soil. There is a hypothesis that plants are able to recruit microorganism by plant exudates, which are composed of amino acids, carbohydrates and organic acids that can vary according to the plant and its biotic or abiotic conditions.46 Different plants select specific microbial communities as reported by Berg et al.,43 when comparing rhizosphere colonization of two medicinal plants: chamomile (Matricaria chamomilla) and nightshade (Solanum distichum), despite being cultivated under similar conditions, they presented different structural (analyzing 16S rRNA genes) and functional (analyzing nitrogen fixing – nifH genes) microbial community. Moreover, plant exudate of the same plant varies according to plant developmental stages selecting specific microbial communities.47 Researchers already identified some plant exudate compounds responsible for specific interactions such as flavonoids in Legume-Rhizobia48 and Strigolactone as a signal molecule for arbuscular mycorrhizal fungi (AMF).49
Reinhold-Hurek et al.,50 proposed a model for microorganism colonization. In bulk soil, the microbial community presents a great diversity and is influenced only by soil type and environmental factors. Getting closer to plant roots (rhizosphere), where there are root exudates, there are fewer species and a more specialized community. And only a few species are able to enter plant root and establish in the plant. Furthermore, after entering the plant, microbial community varies among the different organs: top leaves, fruits, bottom leaves, flowers, stems and roots.51
Mutualistic microorganisms can protect plants from pathogen either by inducing plant resistance or by antibiosis. The induced systemic resistance (ISR) in plants leads to high tolerance to pathogens. There are soils that even if there is the pathogen the disease does not occur, the mechanisms of these disease-suppressive are still being investigated. In this way, Mendes et al.,52 analyzed the microbiome of a soil suppressive to the fungal pathogen Rhizoctonia solani that causes damping off in several agricultural crops. Using a 16S rDNA oligonucleotide microarray (PhyloChip) they were able to identify more than 33,000 bacterial and archaeal taxa in the sugar beet seedlings rhizosphere grown in suppressive soil and in conductive soils. These analyses revealed the bacterial groups present only in the suppressive soil. The authors reported that γ-Proteobacteria, especially Pseudomonadaceae, were all more abundant in suppressive soil than in conducive soil, focusing thereby in this bacterial group. Using random transposon mutagenesis technic in Pseudomonas sp. they were able to identified genes responsible for the biosynthesis of an antifungal: nine-amino acid chlorinated lipopeptide produced by Pseudomonas sp. and controls the pathogen.
From the same PhyloChip diversity analysis, Cordovez et al.,53 identified other antifungal, this time produced by rhizosphere-associated streptomycetes. Theses Streptomyces isolates were able to produce chemically diverse volatile organic compounds (VOCs) with an antifungal activity as well as plant growth-promoting properties. Showing that different bacteria groups can have similar roles in the same environment. Another example was reported by Ardanov et al.,54 who showed that the inoculation of Methylobacterium strains also protected plants against pathogen attack and affected endophyte communities. Therefore, using this concept, researchers started inoculating plants with a pool of microorganism with complementary traits, for example with different mechanisms of control, however, it is a challenge to find the right players to be inoculated.25
In order to define which microorganisms should be inoculated several approaches were used. The first approach seeks to define a core microbiome of a healthy host, or understand the function of microbiomes by sequencing approach, that can be followed by experiments on gnotobiotic host manipulating the microbiome with a selection factor (for example antibiotics, salinity, and U.V. light) or transferring microbiomes between hosts.55
In this way, researchers are starting to study “microbiome engineering”, modulating microbial community. This modulation can occur either by performing plant breeding programs selecting a beneficial interaction between plant lines and rhizosphere microbiome or by redirect rhizosphere microbiome by stimulating or introducing beneficial microorganisms.25,55 The microbiome engineering can occur by altering ecological processes such as modulation in community diversity and structure changing microbe–interaction networks and by altering the evolutionary processes which include extinction of microbial species in the microbiome, horizontal gene transfer, and mutations that can restructure microbial genomes.55
Summarizing, plant phenotype is the sum of plant response to the environment and to the present microbiome (including endophytes and pathogens), this microbiome also responds to the environment and interacts with each other.26 Mendes and Raaijmakers44 suggest a similarity between gut and plant rhizosphere microbiomes. They are both open systems, with a gradient of oxygen, water, and pH resulting in a large number and diversity of microorganism due to the different existing conditions. There are differences between gut and plant rhizosphere microbiome composition, therefore there are some similarities related to nutrient acquisition, immune system modulation and protection against infections. Berg et al.,56 point seven the similarities between host-associated microbiome ecology, among them: different abiotic conditions shape the structure of microbial communities; host and its microbiome co-evolute; core microbiome can be transmitted vertically; during life cycle the microbiome structure varies; host-associated microbiomes are composed of bacteria, archaea, and eukaryotic microorganisms; functional diversity is key in a microbiome; microbial diversity is lost by Human interventions.
Secondary metabolismMicroorganisms produce a large variety of compounds known as secondary metabolites that do not play an essential role in growth, development, and reproduction of the producing organism.57 Nevertheless, these metabolites are often bioactive compounds and can perform important functions in defense, competition, signaling, and ecological interactions.58,59
To establish a microbial interaction network, microorganisms usually respond by metabolic exchange, which leads to complex regulatory responses involving the biosynthesis of secondary metabolites. These interactions can be parasitic, antagonistic, or competitive and the metabolites involved and their functions have been specially studied recently as a result of the advent of tools such as metabolomics and imaging mass spectrometry (IMS) technology.17,60,61
Siderophores are related to competitive and cooperative microbial interactions and can also play other roles, such as signaling and antibiotic activity.62 Hopanoids play an important role in bacterial interaction, conferring tolerance and improving the adaptation of bacteria in different environments.63–65 In fungi, the compounds differentially regulated in an interaction are often bioactive secondary metabolites, such as diketopiperazines, trichothecenes, atranones, and polyketides.14,17 Nevertheless, there is still a lot to understand about the mechanisms involved and the role of many secondary metabolites and genes differentially expressed during the interaction. In this section, we present examples of studies on secondary metabolites involved in different types of microbial interactions.
Endophyte–phytopathogen-plant interactionThe metabolites and mechanisms involved in the interactions between endophyte and phytopathogen and host plant are still very unclear and are predicted to involve many secondary metabolites. Endophytic fungi are known to produce a large variety of bioactive secondary metabolites66,67 that are probably related to the endophyte complex interactions with the host and the phytopathogens and can perform important ecological functions, for example, in the plant development (as growth promoters) and in defense, acting against phytopathogens.68,69
This interaction has been studied in co-cultures of the phytopathogen Moniliophthora roreri and the endophyte Trichoderma harzianum that cohabit in cacao plants.61T. harzianum is extensively used as a biocontrol agent and has known ability to antagonize M. roreri. They identified four secondary metabolites (T39 butenolide, harzianolide, sorbicillinol, and an unknown substance) which production was dependent on the phytopathogen presence and was spatially localized in the interaction zone.61 T39 butenolide and harzianolide have been reported to have antifungal activity. Sorbicillinol is an intermediate in the biosynthesis of bisorbicillinoids, a family of secondary metabolites which present diverse activities.70
Trichoderma atroviride, commonly used as a biocontrol agent, produces acetic acid-related indoles compounds that may stimulate plant growth. Colonization of Arabidopsis roots by T. atroviride promotes growth and enhances systemic disease resistance conferring resistance against hemibiotrophic and necrotrophic phytopathogens.71
Other co-cultured studies were performed with bacteria. Araújo et al.,72 isolated a great number of Methylobacterium strains from asymptomatic citrus plants (with Xylella fastidiosa but without disease), then Lacava et al.,73 showed that Methylobacterium mesophilicum SR1.6/6 and Curtobacterium sp. ER1.6/6 isolated from health and asymptomatic plants inhibited the growth of the phytopathogen Xylella fastidiosa, the causal agent of citrus variegated chlorosis. Moreover, transcriptional profile of Xylella fastidiosa was evaluated during in vitro co-cultivation with a citrus endophytic strain of Methylobacterium mesophilicum. It was shown that genes related to growth, such as genes involved in DNA replication and protein synthesis, were down-regulated. While genes related to energy production, stress, transport, and motility, such as fumarate hydratase, dihydrolipoamide dehydrogenase (Krebs cycle), pilY transporter, clpP peptidase, acriflavin resistance, and toluene tolerance genes, were up-regulated.74
Another approach to study endophyte-phytopathogen-plant interaction is based on the genome sequencing and transposon mutagenesis of an endophyte strain of Burkholderia seminalis, which suppress orchid leaf necrosis by Burkholderia gladioli, revealed eight loci related to biological control. A wcb cluster related to the synthesis of extracellular polysaccharides of the bacterial capsule was identified.75 Extracellular polysaccharides are known to be key factors in bacterial–host interactions.76,77 In addition, genes clusters putatively related to indole-acetic acid and ethylene biosynthesis were identified in the sequenced genome of the endophyte strain, suggesting that this strain might interact with the plant by altering hormone metabolism.75
HopanoidHopanoids compose the cell membrane of some bacteria,78 presenting the same function of eukaryotes cholesterol. They are responsible for stabilization of the membrane and regulates its fluidity and permeability.79 Experiments that knockout biosynthesis genes such as hnpF (squalene hopene cyclase-shc) gene shows that the absence of hopanoids does not influence bacterial growth,79,80 but affects tolerance to several stress conditions, such as extremely acidic environments81; toxic compounds as dichloromethane (DCM)82; it also affects the resistance to antibiotics64 and antimicrobial lipopeptide63; playing a role in multidrug transport83 and bacterial motility.84
Hopanoids act increasing bacteria tolerance to adverse environments, conferring resistance to stress conditions including extreme pH and temperature, and exposure to detergents and antibiotics.63,64 In this way, hopanoids may be involved in bacteria–plant interaction, being responsible for adaptation of bacteria in aerobic micro-environment and low pH culture medium65; as well as involved in nitrogen metabolism in Frankia sp.85 For example, a type of hopanoids produced by the nitrogen-fixing bacteria Bradyrhizobium diazoefficiens is essential for its symbiosis with the host Aeschynomene afraspera, a tropical legume. In this case, the synthesis of C35 hopanoids is related to evasion of plant defense, utilization of host photosynthates, and nitrogen fixation.86
Parasitic interactionThe study of the mycoparasitic interaction between Stachybotrys elegans and Rhizoctonia solani revealed many secondary metabolites differentially expressed in the interaction.17 During the interaction, S. elegans produces cell wall degrading enzymes and express genes associated with parasitism87,88 while R. solani responds with an elevated level of the pyridoxal reductase-encoding gene.89 A metabolomic study showed the profile of the induced secondary metabolites during the interaction. It was showed a significant effect of the mycoparasite on R. solani metabolism, the biosynthesis of many antimicrobial compounds were down-regulated, possibly as a result of the interaction, and only a few diketopiperazines were induced.17 Diketopiperazines are known to have antimicrobial properties, among others biological activities.90 The mycoparasite S. elegans produced several mycotoxins, mainly trichothecenes and atranones. They hypothesized that the trichothecenes were triggered by R. solani and were responsible for the alteration in its metabolism, growth, and development.17 Trichothecenes are a major class of mycotoxins and have been reported to inhibit eukaryotic protein biosynthesis and generate oxidative stress.91
Microbial communities interactionActinomycetes are noteworthy as producers of many natural products with a wide range of bioactivities.60 A study on Streptomyces coelicolor interacting with other actinomycetes showed that most of the compounds produced in each interaction were unique, revealing a differential response in each case. Many unknown molecules and an extended family of acyl-desferrioxamine siderophores never described before in S. coelicolor were identified. They identified 227 compounds differentially produced in interactions; half of these were known metabolites: prodiginines, actinorhodins, coelichelins, and acyl-desferrioxamines. Thus, actinomycetes interspecies interaction seems to be very specific and complex.92
It has been shown that fungal–bacterial interactions can lead to the production of specific fungal secondary metabolites and not only diffusible compounds act in this communication, but there is also a contribution from physical interaction.14 Schroeckh et al.,14 demonstrated that an intimate physical interaction between Aspergillus nidulans and the actinomycete Streptomyces rapamycinicus leads to the activation of fungal secondary metabolite genes related to the production of aromatic polyketides, which were otherwise silent. A PKS gene required for the biosynthesis of the archetypal polyketide orsellinic acid, lecanoric acid (typical lichen metabolite), and the compounds F-9775A and F-9775B (cathepsin K inhibitors) was identified.14 It was later reported that alterations in fungal histone acetylation via the Saga/Ada complex are triggered by the actinomycete leading to the induction of the otherwise silent PKS cluster. This result shows that bacteria can trigger alterations of histone acetylation in fungi.93
SiderophoreThe production and acquisition of siderophores by microorganisms is a crucial mechanism to obtain iron. Many microorganisms secrete siderophores in the environment that when loaded are recognized by cell surface receptors and then transported into the microbial cell.94 Thus, they are related to competitive and cooperative microbial interactions. In addition, many siderophores can also present other functions, for example, they can function as sequesters of a variety of metals and even heavy metal toxins, as signaling molecules, as agents in regulating oxidative stress, and as antibiotics, which were reviewed by Johnstone and Nolan.62
In some Pseudomonas species, a group of siderophores called pyoverdines is essential to infection and biofilm formation, probably helping to regulate bacterial growth.95 Pyoverdines have been reported to act as signaling molecules triggering a cascade that results in the production of several virulence factors, such as exotoxin A, PrpL endoprotease, and pyoverdine itself.96
In the marine environment, exogenous siderophores affect the synthesis of induced siderophores and other iron acquisition mechanisms by others microbial species, working as signaling compounds that influence the growth of marine bacteria under iron-limited conditions. Many strains of marine bacteria were reported to produce siderophores and iron-regulated outer membrane proteins only in the presence of exogenous siderophores produced by other species, such as N,N′-bis (2,3-dihydroxybenzoyl)-O-serylserine from a Vibrio sp., even under very low iron concentrations.97
Symbiotic interactionA remarkably complex inter-kingdom interaction is the symbiotic relationship between Burkholderia, a genus of bacteria, and Rhizopus, a genus of phytopathogen fungi that causes causing rice seedling blight. The endosymbiotic bacteria Burkholderia spp. is responsible for the production of the phytotoxin rhizoxin, the causal agent of rice seedling blight.98 It was reported that in the absence of the endosymbiont, Rhizopus is not capable of produce spores, indicating that the fungus is dependent on factors produced by the symbiont to complete its life cycle.99 This complex symbiont–pathogen–plant interaction is still poorly understood regarding the metabolites and mechanisms involved in the communication and interaction. A study on exopolysaccharide (EPS), which usually plays key roles in interactions, produced by Burkholderia rhizoxinica described a previously unknown structure of EPS. However, the loss of EPS production did not affect the endosymbiotinc interaction with Rhizopus microsporus, as shown by a targeted knockout mutant experiment.100Burkholderia gladioli produces enacyloxins (polyketides with potent antibiotic activity) in co-culture with R. microsporus. The fungus induces the growth of B. gladioli resulting in an increased production of bongkrekic acid, which inhibited the growth of the fungus.101
Quorum sensingQuorum sensing (QS) is the bacterial cell-cell communication. This process involves the production and detection of signaling molecules (called autoinducers) allowing bacterial communities to express genes collectively.102 QS systems are different in Gram-negatives and Gram-positives, the signaling molecules are called acyl-homoserine-lactones (AHLs) in proteobacteria or cis-11-methyl-2-dodecanoic acid (also called diffusible signal factor – DSF) mainly in Xanthomonas and Xylella, gram negatives and gamma-butyrolactones in Streptomyces and peptides in Gram positives.103,104
The first QS system described was in the 1980s in Vibrio fischeri (formerly known as Photobacterium fischeri) bacterium. In the sea, it is in a low population density and does not luminesce. Therefore, when it is in a symbiotic association with fishes and squids it luminesces. After the autoinducer molecule reaches a threshold luminescence genes are activated. This light matches the moonlight, making the squid invisible to the predators below.105,106
In Gram-negative bacteria, two proteins are involved in QS system: the transcriptional regulator R (or LuxR) and the autoinducer synthase I (or LuxI). In V. fischeri this system is called LuxR/LuxI. The signaling molecule or autoinducer (AHL) ligates to the transcriptional regulator LuxR, this ligation is very specific, used for interspecies communication.107,108 Therefore, there are several reports of intraspecific communication as well.109 Some bacteria present more than one system, for example, Rhizobium leguminosarum with five R proteins,110 used for different functions: nodulation efficiency, growth inhibition, nitrogen fixation and plasmid transfer.111
Thereby, QS can have different roles: fluorescence emission (as reported above with Vibrio fischeri example), virulence, sporulation, competence, antibiotic production and biofilm formation,102 and can act during the interaction of different organism: bacteria–bacteria, fungal–bacteria, bacteria–host (animal or plant). It regulates a large number of genes, around 6–10% of the microbial genome.103
In gram-positive bacteria, specifically Bacillus subtilis and Streptococcus pneumoniae peptide signal can induce sporulation and competence development. This was evidenced by experiments showing that sporulation and competence are inefficient at low cell densities and needs a secreted bacterial factor.112
Concerning virulence, pathogens are able to control virulence factors expression by QS molecule. Vascular pathogen, such as Xanthomonas and Xylella uses DSF signaling to express virulence factor as well as biofilm formation104Xylella also uses DSF signaling to colonize the insect vector, which is key in the disease transmission.113 Other vascular pathogen Pantoea stewartii uses AHL molecules to express disease, QS mutants of P. stewartii were not able to disperse and migrate in the vessels, consequently decreasing the disease.114 The epiphytic plant pathogen Pseudomonas syringae also uses AHL molecule in virulence. This bacterium is able to control motility and exopolysaccharide synthesis essential on biofilm formation and leave colonization.115 Therefore, QS inhibitors (QSI) can reduce biofilm formation and increase de bacterial susceptibility to antibiotics. There are four strategies used to interfere with QS inhibition of: 1. signal generation; 2. signal dissemination, 3. Signal receptor and signaling response system.116,117
Reports of AHL degradation by environmental and clinic bacteria, affecting AHL signaling have been described. For example, P. aeruginosa and Burkholderia cepacia are associated to pneumoniae in cystic fibrosis patients and during this infection the cross talk seems to be an important strategy for both bacteria. P. aeruginosa produces AHL able to induce B. cepacia genes involved in biofilm formation.111 On the other hand, Non AHL producing bacteria can foreclose AHL movement, affecting AHL – mediated responses.118 This cross-talking can occur between other organisms such as plants and bacteria, which is key during plant–bacteria interaction. Plants produce compounds that mimics AHL and interferes with AHL biosensors,119 for example, Medicago sativa may produce a compound able to inhibit exopolysaccharides production in Sinorhizobium meliloti.120 QS also regulates conjugative transfer during plant-Agrobacterium tumefaciens interaction, which bacteria induce crown-gall by transferring T-DNA, that codifies proteins involved in opine biosynthesis, to the plant. The conjugation is trigged by AHL molecules.111 This cross-talking can also occur bacteria–fungus and bacteria–animal. Fungus and animals can produce compounds that inhibit QS-controlled genes in P. aeruginosa.116,121,122
The Candida albicans and Pseudomonas aeruginosa interaction is an important model that show how fungi and bacteria can regulate each other by QS system. Farnesol (a sesquiterpene) and tyrosol's produced by Candida albicans are associated to control the physiology and virulence of this fungi. In fact, farnesol is associated to resistance to drugs, antimicrobial activity and inhibition of filamentation stage and biofilm formation, while tyrosol induces oxidative stress resistance, a shortened lag phase of growth and stimulate the germ tube in yeast cells and hyphae in the early stage of biofilm formation.123 In the host, Candida albicans may share the same environment with the bacterium P. aeruginosa, which bacterium may present a complex QS system based on the synthesis of many molecules, such as 3-oxo-C12 homoserine-lactones (HSL) and 2-heptyl-3-hydroxy-4-quinolone (PQS–Pseudomonas quinolone signal). P. aeruginosa may attach on to a filamentous form of C. albicans and inhibit this fungus by synthesizing many molecules, including phenazines, pyocianyn, haemolytic phospholipase C,124 suggesting that these molecules are associated with niche construction during establishment in the host. During this interaction, the P. aeruginosa QS system may block the yeast-to-hypha transition or activates the hypha-to-yeast reversion, suggesting that C. albicans may sense the presence of the bacterium and activates a survival mechanism.123 In another hand, the farnesol produced by C. albicans downregulate the PQS system of P. aeruginosa, inhibiting, in turn, the pyocyanin production.125 This cross-talk between P. aeruginosa and C. albicans and based on the synthesis of farnesol, HSL and PQS allow the coexistence of these microbes in the same environment and control the population level of both, showing that this system may regulate the multi-trophic interaction in complex communities.
Concluding remarksIn the environment, microorganisms live in close contact with many different hosts and with each other in communities, usually including many species. In addition, they are also exposed to variation in the environmental conditions, which in turn affect the interaction among microorganisms and the host. The studies in microbial ecology, including the interaction among microbial species and between microorganism and the host has led to important findings in the ecology, human healthy and biotechnological researches, such as molecular mechanisms related to physiological response in human systemic diseases and antimicrobial drug development based on natural products, synthetic biology and quorum sensing.
Microbial interactions are highly complex and many mechanisms and molecules are involved, enabling that some microorganisms identify some species and respond to each other in a complex environment, including shifts in physical-chemical condition and presence of different hosts, many of them were presented in this review. However, there is still a lot to understand about the “molecular language” used by microorganisms and the molecules and signs related to interaction with the host. The development and adaptation of tools and methods including in vitro and in vivo models are still highly required to better understand and characterize the microbial interactions with more molecular details. In addition, understanding the connection between genomes, gene expression, and molecules in complex environments and communities comprise a very difficult challenge. The ways in which microbial species interact with each other and with the host are a complex issue that is only beginning to be understood, but recent studies have provided new insights in microbial interactions and their application in ecology and human healthy.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by a grant from the Foundation for Research Assistance, São Paulo State, Brazil (Proc. 2012/24217-6 and 2015/11563-1). We thank FAPESP for M.N.D. (Proc. 2013/17314-08) and CNPq for R.M.B. (Proc. 141145/2012-9) fellowships. W.L.A. received Productivity-in-Research fellowship (Produtividade em Pesquisa – PQ) from the National Council for Scientific and Technological Development (CNPq).