Acinetobacter baumannii is widely recognized as an important pathogen associated with nosocomial infections. The treatment of these infections is often difficult due to the acquisition of resistance genes. A. baumannii presents a high genetic plasticity which allows the accumulation of these resistance determinants leading to multidrug resistance. It is highlighted the importance of the horizontal transfer of resistance genes, through mobile genetic elements and its relationship with increased incidence of multidrug resistant A. baumannii in hospitals. Considering that resistance to carbapenems is very important from the clinical and epidemiological point of view, the aim of this article is to present an overview of the current knowledge about genetic elements related to carbapenem resistance in A. baumannii such as integrons, transposons, resistance islands and insertion sequences.
The Acinetobacter baumannii-calcoaceticus (Abc) complex has emerged as an important nosocomial pathogen. Among the members of this complex, A. baumannii, A. pittii, and A. nosocomialis are the three most common Acinetobacter species isolated in clinical settings.1A. baumannii has been extensively studied due to its association with infections of high mortality rates. A. pittii and A. nosocomialis are increasingly identified as causative agents of nosocomial infections.2
A. baumannii is considered an important nosocomial pathogen, causing a wide range of infections, including ventilator-associated pneumonia, bloodstream infections, urinary tract infections and meningitis. This species is naturally highly resistant to a number of antimicrobials commonly used in the clinical practice, such as first and second generation cephalosporins, aminopenicillins, and chloramphenicol. A. baumannii contains an intrinsic AmpC β-lactamase (blaADC) and OXA-51 serine-type oxacilinase (blaOXA-51), which contribute to the natural resistance to β-lactams.3 Moreover, this organism presents a great capacity to acquire new resistance mechanisms, including those responsible for carbapenem resistance.4
Carbapenem resistance in A. baumannii involves mainly the carbapenem-hydrolysing class D β-lactamases (CHDLs – Ambler class D) and less frequently, the metallo-β-lactamases (MBLs – Ambler class B). Carbapenem resistance may also be caused by other mechanisms such as, production of other carbapenemases, porin modification or loss, or by modification of the penicillin-binding proteins.1,5
Several acquired class D OXA-type β-lactamases have been identified as a source of carbapenem resistance in A. baumannii. Five main groups of CHDLs have been described in A. baumannii, corresponding to OXA-23-like, OXA-24/40-like, OXA-58-like, OXA-143-like and OXA-235-like enzymes.6 OXA-23-like enzymes are the most widespread in A. baumannii worldwide and have been identified in all continents.6
In Brazil, OXA-23-like-producing A. baumannii is disseminated in many states and it is responsible for high endemic levels of multidrug-resistance.7,8 The blaOXA-143 gene has thus far been detected only in A. baumannii isolates from Brazil and is the second most frequent CHDL encoding gene.9–11
The blaOXA-143 gene is frequently found in the Southeast region of Brazil, especially in the state of São Paulo. It is important to note that two new variants of this gene were recently described. The variants blaOXA-235 and blaOXA-231 were described in Minas Gerais and Paraná states, respectively.12,13 This data demonstrates the detection of these new variants of blaOXA-143 in Brazil is a cause of great concern and shows the potential of these new CHDLs to spread to other Brazilian regions.
Although blaOXA-24/40-like gene is disseminated in A. baumannii in Europe, in Brazil, this gene is still rare, with only a very few reports of a blaOXA-72 (blaOXA-24/40-like variant) in São Paulo,9 Recife,14 Porto Alegre and Curitiba.
Despite MBLs are less commonly identified in A. baumannii than the OXA-type carbapenemases, their hydrolytic activities to carbapenems are significantly more potent. Four MBLs have been identified in A. baumannii: IMP, VIM, SIM and, more recently, NDM.15 It is important to note that MBL genes, such as NDM and IMP-1, have been described in Acinetobacter non-baumannii species, which demonstrates the capacity of these resistance genes to spread among different Acinetobacter species.16,17
Most of Ambler class A ESBLs possess activity against penicillins and broad-spectrum cephalosporins. However, specific GES variants have been shown to possess the ability to compromise the efficacy of carbapenems. Among A. baumannii, the variants GES-11 and GES-14 possess specific residues enlarging their hydrolysis spectrum (Table 1).18,19
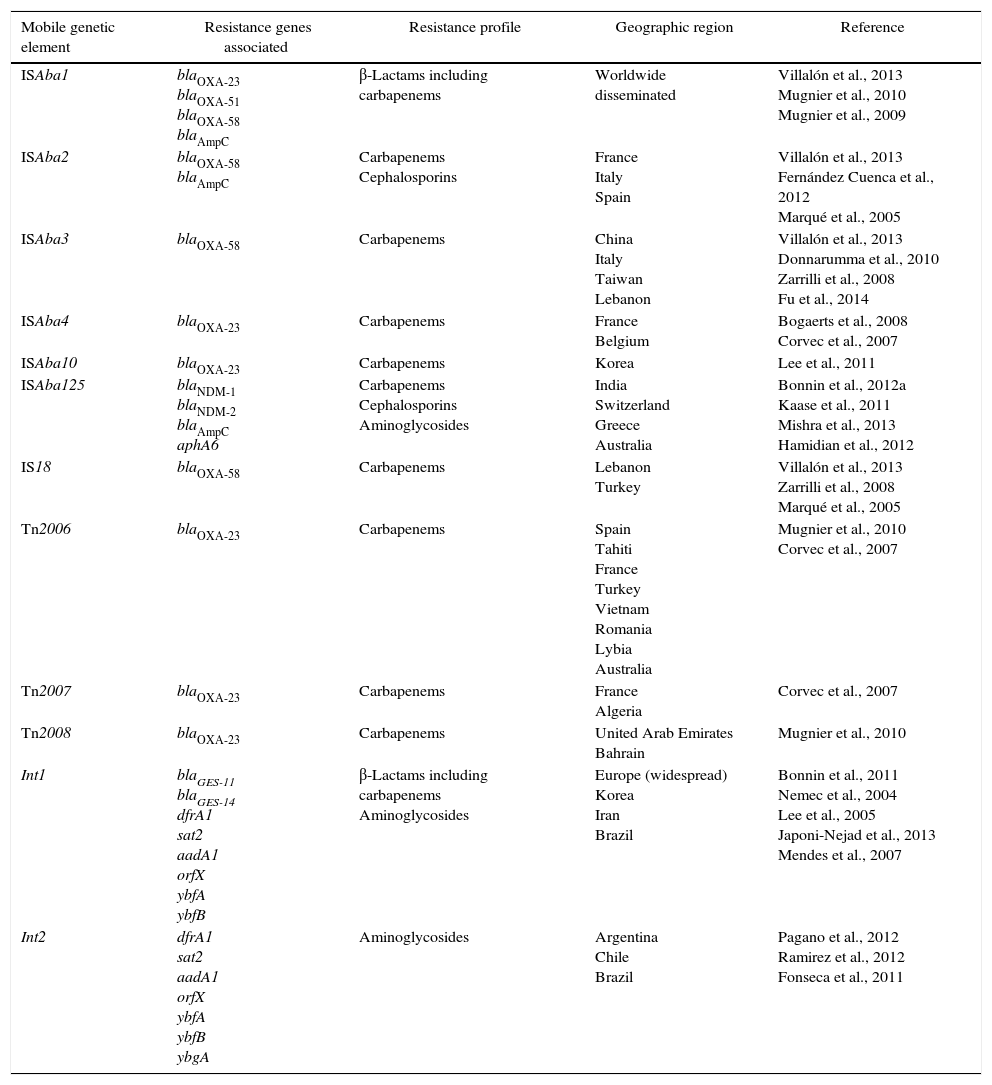
Characterization of the main mobile genetic elements associated with resistance in Acinetobacter baumannii.
| Mobile genetic element | Resistance genes associated | Resistance profile | Geographic region | Reference |
|---|---|---|---|---|
| ISAba1 | blaOXA-23 blaOXA-51 blaOXA-58 blaAmpC | β-Lactams including carbapenems | Worldwide disseminated | Villalón et al., 2013 Mugnier et al., 2010 Mugnier et al., 2009 |
| ISAba2 | blaOXA-58 blaAmpC | Carbapenems Cephalosporins | France Italy Spain | Villalón et al., 2013 Fernández Cuenca et al., 2012 Marqué et al., 2005 |
| ISAba3 | blaOXA-58 | Carbapenems | China Italy Taiwan Lebanon | Villalón et al., 2013 Donnarumma et al., 2010 Zarrilli et al., 2008 Fu et al., 2014 |
| ISAba4 | blaOXA-23 | Carbapenems | France Belgium | Bogaerts et al., 2008 Corvec et al., 2007 |
| ISAba10 | blaOXA-23 | Carbapenems | Korea | Lee et al., 2011 |
| ISAba125 | blaNDM-1 blaNDM-2 blaAmpC aphA6 | Carbapenems Cephalosporins Aminoglycosides | India Switzerland Greece Australia | Bonnin et al., 2012a Kaase et al., 2011 Mishra et al., 2013 Hamidian et al., 2012 |
| IS18 | blaOXA-58 | Carbapenems | Lebanon Turkey | Villalón et al., 2013 Zarrilli et al., 2008 Marqué et al., 2005 |
| Tn2006 | blaOXA-23 | Carbapenems | Spain Tahiti France Turkey Vietnam Romania Lybia Australia | Mugnier et al., 2010 Corvec et al., 2007 |
| Tn2007 | blaOXA-23 | Carbapenems | France Algeria | Corvec et al., 2007 |
| Tn2008 | blaOXA-23 | Carbapenems | United Arab Emirates Bahrain | Mugnier et al., 2010 |
| Int1 | blaGES-11 blaGES-14 dfrA1 sat2 aadA1 orfX ybfA ybfB | β-Lactams including carbapenems Aminoglycosides | Europe (widespread) Korea Iran Brazil | Bonnin et al., 2011 Nemec et al., 2004 Lee et al., 2005 Japoni-Nejad et al., 2013 Mendes et al., 2007 |
| Int2 | dfrA1 sat2 aadA1 orfX ybfA ybfB ybgA | Aminoglycosides | Argentina Chile Brazil | Pagano et al., 2012 Ramirez et al., 2012 Fonseca et al., 2011 |
The elevated genetic plasticity presented by A. baumannii has allowed the accumulation of many resistance determinants, which contributed to the high incidence of A. baumannii multirresistant to antibiotics. In this review, we present and discuss the characteristics of the different mobile genetic elements involved in the transfer of resistance determinants in A. baumannii.
AbaR-type genomic resistance islandsGenomic islands containing resistance markers are referred to as resistance islands. Resistance islands have been described mainly in Proteobacteria, including Shigella flexneri, Salmonella enterica, Vibrio cholerae, Staphylococcus aureus, and more recently, in A. baumannii.20,21A. baumannii isolates harbor large clusters of horizontally transferred genes conferring resistance to multiple antibiotics and heavy metals, which are integrated at a specific site in a particular ATPase gene.22
Fournier et al. described for the first time the A. baumannii Resistant Island (AbaR). AbaR is defined as a region which has transposed into a specific position in the chromosome, creating a 5bp duplication site (ACCGC).21 The backbone of AbaR is comprised of five open reading frames (ORFs) – orf1, tniA, tniB, orf2, orf3 – which constitute the transposition module, and two other genes encoding to the universal stress protein (uspA) and a sulfate permease (sul).21–23
Several AbaR have already been described containing a variety of resistance genes, including the blaOXA-23-like, which confers resistance to carbapenems.24 These resistance islands have been described in A. baumannii epidemic strains belonging to the important global clones, European Clone I (EC I) and European Clone II (EC II), known for their increased capacity to spread worldwide.22
Several other genomic resistance islands have been fully characterized in A. baumannii. The majority were found in strains of EC I, such as, AbaR1, AbaR3, AbaR5, AbaR6, AbaR7, AbaR8, AbaR9, and AbaR10. These AbaRs share a structure represented by a 16.3kb backbone transposon (Tn6019) interrupted by a large compound transposon that contains a variable-resistance region bounded by directly oriented copies of Tn6018. Exceptions are AbaR6 and AbaR7, each with a large deleted region.25 Much less is known about AbaRs in EC II. The resistance islands harbored by this clone are integrated at the same site of the ATPase gene as is known for AbaRs in EC I.25
AbaR1 is the largest resistance island described to date. This island contains 86kb and was originally described in the epidemic A. baumannii strain AYE belonging to ECI. This strain was responsible for outbreaks in France during 2004.21A. baumannii AYE strain revealed the presence of a large gene cluster, containing many resistance determinants, inserted into the chromosome.21
Of the 45 resistance genes described in AbaR1 resistance island, 25 were associated with resistance to several classes of antibiotics. These include genes that had not been previously described in Acinetobacter species such as strA, strB, aphA1, and aac69 (encoding resistance to aminoglycosides); putative tetracycline-resistance genes tetA (tetracycline efflux pump) and tetR (repressor protein); dfrX (resistance to cotrimoxazole); and the chloramphenicol-resistance gene cmlA (chloramphenicol efflux pump). Moreover, Fournier et al. (2006) described the presence of genes in AbaR1 that encode VEB-1 and OXA-10 β-lactamases, the aminoglycoside acetyltransferase gene aac3, and the aminoglycoside adenyltransferases aadA1/DA1/B; the cotrimoxazole resistance-associated dfrI; cmlA5 and one copy of the chloramphenicol acetyl-transferase cat; the rifampin ADP-ribosyltransferase gene arr-2; and five copies of the sulfonamide-resistance gene sulI encoding dihydropteroate synthetase, a component of class 1 integrons.21,26
AbaR2 was described in the epidemic, multidrug-resistant A. baumannii strain named ACICU.27 This strain belongs to ECII and carries the plasmid-mediated blaOXA-58. A. baumannii AYE and ACICU belong to different clonal groups (European clones I and II, respectively), however, the presence of related resistance islands in both lineages suggests that AbaR1 and AbaR2 derived from an island acquired by a common A. baumannii ancestor before their divergence into two different clonal lineages.21,27
The genomic resistance island variant AbaR3 appears to be an ancestral of several AbaR variants which have arisen from AbaR3 by loss of segments of different lengths that include one or more of the antibiotic resistance genes.28 AbaR3 contains eight genes associated with antibiotic resistance. Unique sequences in AbaR3 include a blaTEM gene that is associated with a Tn3 transposon and a small cluster of genes, including two that encode to a DNA topoisomerase and a single-strand binding protein that are similar to proteins from a broad-host-range plasmid.29 In addition, it is noteworthy that the presence of genes for a plasmid-derived DNA topoisomerase may contribute to the resistance island mobility.
Transposable elementsTransposable elements have the ability to move within the bacterial genome, being able to translocate themselves from one site of the genome to other sites. These transpositions are considered one of the major causes of bacterial DNA rearrangements, which in turn can cause changes in gene expression.30 In A. baumannii, transposable elements, such as transposons and insertion sequences have been responsible for the expression and spread of antimicrobial resistance mechanisms.1
Insertion sequencesBacterial insertion sequences (IS) are the least complex type of transposable elements; they rarely exceed 2kb in size and may be as small as 0.5kb. These elements possess an important role in the spread of resistance genes since the presence of two copies of the same IS flanking a resistance gene form a complex structure called composite transposon. Composite transposons are able to mobilize a variety of resistance genes, contributing to antimicrobial resistance dissemination.31
Besides their transposition role, some IS have been shown to activate or to increase the expression of neighbor genes. This capacity may be due to the presence of promoter regions in the insertion sequence or by the formation of new promoters after the insertion event.31
Some IS elements have an important role in A. baumannii antimicrobial resistance. ISAba1, ISAba2, ISAba3, ISAba4 and IS18 are commonly associated with the expression of carbapenemases genes in A. baumannii (Table 1).32 Villalón et al. (2013) investigated the presence of these IS elements in 59 multidrug-resistant A. baumannii isolates and observed a prevalence of 93.2%, 25.4%, 20.3% and 5.1% for ISAba1, ISAba2, ISAba3 and IS18, respectively. ISAba4 was not detected in any of the isolates in this study.32
It is important to note that IS elements such as ISAba1 can contribute to the spread of carbapenemase genes among different Acinetobacter species. Poirel et al. (2008) hypothesized that blaOXA-23 was likely mobilized by the ISAba1 insertion sequence from A. radioresistens to A. baumannii.33 The authors demonstrated that A. radioresistens is the progenitor of the blaOXA-23-like gene, which was mobilized to A. baumannii through ISAba1 insertions sequence provided by A. baumannii. This hypothesis is based on the identification of genes encoding both OXA-23-like and ATPase-like enzymes on the A. radioresistens chromosome without the presence of ISAba1 elements, that is involved in the mobilization of blaOXA-23 gene.33
The ISAba1 element belongs to the IS4 family and has 11-bp inverted repeats sequences flanked by 9-bp direct repeats of the target sequence. Although this element is considered exclusive to A. baumannii, Segal et al. (2005) identified ISAba1 in Acinetobacter lwoffii isolates, demonstrating the high mobility of these elements and indicating that transposition events of the ISAba1 occur frequently.34
ISAba1 has been found upstream the blaOXA-23-like, blaOXA-51-like, blaOXA-58-like and blaAmpC genes in A. baumannii. This IS acts as a promoter sequence which increases the expression of resistance genes. In fact, it was demonstrated that it is necessary the presence of ISAba1 upstream blaOXA-23 and blaOXA-51 for these genes to confer resistance to carbapenems.35 Although several authors have demonstrated the relationship between ISAba1 upstream blaOXA-51 and carbapenem resistance, this may not be enough to confer resistance, as A. baumannii isolates susceptible to carbapenems with the association ISAba1/blaOXA-51 have already been described.36
The ISAba2, ISAba3 and ISAba4 elements have also been identified upstream blaOXA-58-like and blaOXA-23-like genes in A. baumannii isolates.37 Giannouli et al. (2009) analyzed the insertion sequences of 24 A. baumannii isolates with blaOXA-58 gene and identified the presence ISAba2, IS18 or ISAba1 located at the 5′ end, while at 3′ end all isolates presented the ISAba3 element. Of note, the IS elements at 5′ end of blaOXA-58 were evidenced in strains of distinct PFGE profiles and ST groups in the same geographical area. It suggests that these elements might have been acquired through horizontal gene transfer and confirms their dissemination capacity among A. baumannii isolates.38
Corvec et al. (2007) described the first A. baumannii isolate harboring an ISAba4 element upstream blaOXA-23 gene, in France. Subsequently, it was shown that an isolate from Belgium containing the association of ISAba4 and the blaOXA-23 presented the same PFGE profile as the isolate from France. These findings demonstrate the propensity of resistant strains to spread, highlighting the importance of epidemiological surveys to estimate the true prevalence of isolates harboring ISAba4/blaOXA-23.39,40
Lee et al. (2011) identified a novel 1203bp insertion sequence, named ISAba10. This element was found to be inserted into the ISAba1 element upstream blaOXA-23 gene in an A. baumannii presenting high minimum inhibitory concentrations (MICs) to carbapenems (≥32μg/mL). In addition, isolates without the insertion of this element showed MICs between 8 and 16μg/mL. The authors suggested that this sequence may increase 2–5-fold the blaOXA-23 gene expression. Based on these results, they suggested that the ISAba10 element may play an important role in carbapenem resistance by providing an additional promoter sequence to the blaOXA-23 gene.41
IS elements have also been associated to metallo-β-lactamases such as blaNDM, which have been increasingly reported in Acinetobacter baumannii and in other Acinetobacter species such as A. johnsonnii, A. pittii, A. junii and A. lwoffii.16,42–44 The blaNDM can be located either on the plasmid or chromosome in Acinetobacter species.44 However, it was evidenced that the spread of the blaNDM gene was not associated with clonal dissemination, but horizontal spread of the genetic structure.42
Several studies reported that blaNDM gene is located between two copies of the ISAba125 element, forming a composite transposon named Tn125. ISAba125 element provides the -35 sequence of the hybrid promoter responsible for the expression of the blaNDM gene.45 Curiously, this IS element has been originally identified from an A. baumannii isolate without any association with the blaNDM gene. By contrast, this IS has been identified in Enterobacteriaceae and P. aeruginosa as a remnant of the Tn125 and has never been identified alone in these species. This observation suggests that A. baumannii is a likely reservoir of ISAba125. Findings like these highlight that even though A. baumannii is usually recognized as a final acceptor for resistance genes, it may acquire several resistance determinants and then transfer them to Enterobacteriaceae and Pseudomonas spp.
Recently, a study demonstrated that Tn125 has been disrupted by IS26 in A. baumannii NDM-producing isolates from India. This new rearrangement has resulted in blaNDM-1 being within an IS26 composite transposon, which might potentially mobilize blaNDM-1 and contribute to the spread of the carbapenemase gene.46
Robledo et al. (2010) described the first report of blaKPC gene in A. baumannii isolates from Puerto Rico. In that study, four variants of blaKPC were identified: KPC-2, -3, and -4 and a novel variant, KPC-10. The integration of these genes in the A. baumannii chromosome was related to a transposition event mediated by the transposase of ISEcp1.31,47 This element is likely to be responsible for mobilizing numerous blaCTX-M genes and several other resistance genes such as qnrB19, rmtC, blaACC-1 and blaCMY-2g7,16,21.48 In addition, it was responsible for the mobilization of blaCTX-M-5 from a narrow range plasmid to the chromosome of A. baumannii, event similar to what Martinez et al. observed with blaKPC gene.47
As described above, ISs can cause insertion mutations, genome rearrangements and enhance the spread of resistance and virulence determinants within pathogenic species. Besides being involved in the expression and spread of carbapenemases, IS elements such as ISAba1, ISAba10 and ISAba825 are involved on the disruption of carO gene, which codes for an important outer membrane channel. The absence of this outer membrane protein has been correlated with reduced susceptibility to carbapenems.41,49,50
TransposonsTransposons sequences may vary in size from 3 to 40kb, in some cases containing dozens of genes. These elements are into two main classes: composite transposons or complex transposons. Composite transposons have resistance genes in its central region; furthermore, these elements are flanked by an insertion sequence (IS) at each end. Complex transposons have a more complicated genetic structure than IS elements or composite transposons. The classic complex transposon is Tn3, which is derived from resistance plasmid R1.51
In A. baumannii, transposons have been characterized as genetic structures harboring important resistant genes, such as blaOXA-23. Three transposons have been related to blaOXA-23: Tn2006, Tn2007 and Tn2008. In Tn2006, the blaOXA-23 gene is flanked by two copies of the insertion sequence ISAba1, which is located in opposite directions. Tn2008 is similar to Tn2006 but lacks the second copy of ISAba1. Finally, in Tn2007 the blaOXA-23 gene is associated with one copy of ISAba4 located upstream to this gene.52 Several studies have demonstrated that Tn2006 is currently the most common determinant of carbapenem resistance, with a great ability to spread among A. baumannii isolates.53
IntegronsThese elements are natural cloning and expression systems that incorporate ORFs by site-specific recombination and convert them to functional genes due to the presence of a promoter sequence (Rowe-Magnus et al., 2001). It is now well established that these mobile elements constitute the major vectors of antibiotic multiresistance in Gram-negative and, to a lesser extent, in Gram-positive bacteria.54
Five different classes of mobile integrons have been defined to date, based on the sequence of the encoded integrases.54,55 It is known that three (classes 1, 2 and 3) of these classes have an important role in the dissemination of antimicrobial resistance genes.55,56 These classes are well described in the literature and are associated to multiresistant phenotypes.54,55
Several studies have demonstrated a high prevalence of class 1 integrons in A. baumannii isolates in Europe, Asia and United States.57 Due to its greater spread capacity, class 1 integrons are the main experimental model of integrons. This class is usually associated to functional or non functional transposons derived from Tn402 which may be inserted into larger transposons as Tn21. Class 1 integrons have been associated to a variety of insertion sequences, including IS26, IS1999, IS2000 e IS6100.58
Most acquired MBL genes in A. baumannii have been found within class 1 integrons, often containing an array of resistance gene cassettes.1,6 Mendes et al. (2007) described seven blaIMP-1 harboring Acinetobacter spp. isolates recovered from Brazilian inpatients. All isolates possessed a class 1 integron, named In86, carrying the same cassette array: blaIMP, aac(6′)-31, and aadA1, which was plasmid-located in five of the isolates (Mendes et al., 2007). This gene cassette contained a aminoglycoside resistance gene – aac(6′)-31 – that might be capable of conferring resistance to all clinically available aminoglycosides. This gene was able to disseminate among unrelated A. baumannii clinical isolates from a Brazilian hospital (Mendes et al., 2007). Recently, Cayô et al., reported a new structure of class 1 integron, In990, harboring the blaIMP-10 in A. baumannii isolates from Brazil. The cassette arrangement of In990 was very similar to that of In86 described by Mendes et al.59,60
Class 2 integrons are included in the Tn7 family of transposons, and consist of an integrase gene followed by gene cassettes. Tn7 are identified as a sophisticated mobile genetic element containing a transposition module formed by five transposition genes, tnsA, tnsB, tnsC, tnsD, and tnsE, rather than the one or two seen in many other transposable elements.56 Class 3 integrons are less prevalent than class 2 and are also located in transposons.54
Despite reports of a higher prevalence of class 1 integrons in A. baumannii, studies conducted in Latin American countries such as Chile, Argentina and Brazil demonstrated a greater distribution of class 2 integrons among isolates of A. baumannii in these regions.36,61 Fonseca et al. (2011) demonstrated that all class 2 integrons obtained from Brazilian isolates were inserted into Tn7 transposon, besides having the gene cassette containing the arrangement of genes dfrA1 (trimethoprim resistance), sat2 (streptothricin resistance) and aadA1 (spectinomycin and streptomycin resistance).62
Martins et al. (2015) investigated the association of class 2 integrons and gene cassettes with clonal lineages of A. baumannii. They reported the association of class 1 and 2 integrons with CC109/1 (International Clone I) and CC113/79 A. baumannii strains, respectively. The authors hypothesized that class 2 integron, predominant in Latin America, may be accounted for the high prevalence of the CC113/79 type. In the same study, a similar prevalence was observed for A. nosocomialis.63
Class 1 and 2 integrons have been described in A. baumannii isolates related to nosocomial infection outbreaks. In a study published by Turton et al. (2005), it was observed that all A. baumannii isolates associated with outbreaks contained class 1 integrons, in contrast, none sporadic isolate presented this class of integron.64
More than 130 different gene cassettes containing resistance genes have been identified in integrons. Distinct genes are evidenced in gene cassettes, promoting resistance to a variety of antimicrobial classes. Together, these gene cassettes provide resistance to most classes of antibiotics including β-lactams, all aminoglycosides, chloramphenicol, trimethoprim, streptothricin, rifampin, erythromycin, fosfomycin, lincomycin, quinolones, and antiseptics of the quaternary ammonium-compound family.65 Besides these genes, several ORFs with unknown function have been identified in gene cassettes.66
In A. baumannii, gene cassettes have been described containing several genes, such as aacA4 responsible for resistance to amikacin, netilmicin and tobramycin, the catB8 gene is an acetyltransferase which encodes resistance to chloramphenicol, aadA1 is responsible for resistance to streptomycin and spectinomycin, aac3 responsible for resistance to gentamicin and blaOXA-10 encodes resistance to β-lactams, except carbapenems and extended-spectrum cephalosporins.21
Final remarksThis review highlighted the role of resistance determinants in the capacity of spread in A. baumannii. This species shows a considerable ability to acquire foreign DNA such as drug resistance genes, which provide a genetic diversity and overcomes the antibiotic selection pressure. It is important to note that the main carbapenem-resistance mechanism involved in A. baumannii (production of oxacillinases) presents a low hydrolytic power when it is not associated with an insertion sequence. Moreover, the capacity of OXA genes to spread is directly related to their association with a composite transposon (Tn2006). These features highlight the importance of investigating the genetic context of these genes in order to define their real clinical significance.
The continuous description of gene cassettes in integrons, mainly those leading to resistance to β-lactams and aminoglycosides, has been of great concern. Furthermore, the number of resistance genes inserted in the same plasmid, even in the same integron, seems to be increasing. This integration of resistance determinants in the same plasmid may facilitate the persistence in the environment for long periods because of the physical association of integrons with other elements, allowing their continued selection.57
In this context, the knowledge about the genetic structure of resistance determinants is very important in order to understand the capacity of resistance genes to spread in A. baumannii.
Conflicts of interestThe authors declare no conflicts of interest.