The aim of this study was to perform the molecular characterization of conserved and variable regions of feline calicivirus capsid genome in order to investigate the molecular diversity of variants in Brazilian cat population. Twenty-six conjunctival samples from cats living in five public short-term animal shelters and three multicat life-long households were analyzed. Fifteen cats had conjunctivitis, three had oral ulceration, eight had respiratory signs (cough, sneeze and nasal discharge) and nine were asymptomatic. Feline calicivirus were isolated in CRFK cells and characterized by reverse transcription PCR target to both conserved and variable regions of open reading frame 2. The amplicons obtained were sequenced. A phylogenetic analysis along with most of the prototypes available in GenBank database and an amino acid analysis were performed. Phylogenetic analysis based on both conserved and variable region revealed two clusters with an aLTR value of 1.00 and 0.98 respectively and the variants from this study belong to feline calicivirus genogroup I. No association between geographical distribution and/or clinical signs and clustering in phylogenetic tree was observed. The variants circulating in public short-term animal shelter demonstrated a high variability because of the relatively rapid turnover of carrier cats constantly introduced of multiple viruses into this location over time.
Feline calicivirus (FCV) belongs to the family Caliciviridae, genus Vesivirus, and is usually associated with moderate, self-limiting acute oral and upper respiratory tract disease (URTD), chronic stomatitis and oral ulceration.1 FCV can also cause conjunctivitis, limping syndrome and a severe clinical form known as FCV-associated virulent systemic disease (FCV-VSD).2–6
The genome of FCV is a small single-stranded, positive-sense RNA of approximately 7.7kb that contains three open reading frames (ORFs). ORF 1 encodes the nonstructural proteins. ORF 2 is divided into six regions A-F. The genomic regions A, B, D and F are relatively well conserved among FCV, while regions C and E are variables with significant sequence divergence. Region E still is subdivided into 5′ and 3′ hypervariable regions (HVRs) separated by a conserved central domain (conE).7,8 ORF 2 encodes the major capsid protein (VP1) and ORF 3 encodes the virus minor structural protein (VP2).9 The 5′HVR encodes the major B-cell epitopes being the target VP1 region for virus-neutralizing antibodies10 and the high variability of the region E is the basis for molecular characterization and classification of different FCV strains.1,11
FCV genetic diversity leads to quasispecies virus evolution, important to the emergence of strains causing a severe and acute virulent systemic disease and also affords virus adaptability to new environmental niches.8,9 Most vaccines are based on the broadly cross-reactive F9 attenuated strain or 255 inactivated strain,12 nevertheless vaccine does not prevent reinfection by homologous or heterologous FCV variants.13,14 So, the prevalence of FCV in the general cat population, especially among colonies or shelters remains high.15
Little is known about the molecular diversity of FCV variants in cats from shelters in Brazil. The aim of this study was to perform FCV molecular characterization directed to conserved and variable regions of FCV capsid (A-B and C-F) isolated from shelters and household cats from Rio de Janeiro, Brazil.
Material and methodClinical specimensTwenty-six conjunctival samples were collected from cats up to one year-old living in three multicat life-long households and five public short-term animal shelters in Rio de Janeiro, Brazil, from 2013 to 2015. The mean geographical distance among the collection sites was 68.8km including two cities (Rio de Janeiro and Niterói). In this study, samples previously tested positive for FCV by nested RT-PCR16 were used too.
The multicat life-long household was characterized as a house with more than 10 vaccinated or unvaccinated cats living in the dwelling for a long period of time without outside access and with fomite sharing. The public short-term animal shelter received cats from many different places and they were grouped in cages for a short period of time with veterinary care. Cats were unvaccinated. There were dogs living in the public shelters, but they did not have contact with cats. Both places commonly lacked of a minimal standard of hygiene.
Informations about age, sex, clinical signs of URTD, vaccination status and local (if multicat household or public shelter) were recorded on an individual medical record. This study was approved by Institutional committee on animal welfare and ethics (UFF – approval number 708/2016).
Cell and virus isolationCrandell-Rees Feline kidney cells used for virus isolation were cultured in Dubelcco's modified Eagle's minimal essential medium (DMEM). Animal specimens from the conjunctival sacs were taken rolling a cytobrush (Cytobrush PlusR, Trumbull) in the ventral conjunctival fornix17 and pooled by immersing and shaking in a 2.0mL microtube containing 0.5mL of cell culture DMEM plus antibiotics. All samples were filtered and stored to −70°C. For virus isolation, 100μL samples were inoculated onto CRFK cells monolayers and incubated at 37°C in a CO2 incubator. Monitoring for appearance of cytopathic effect (CPE) was performed for four days. All homogenates were submitted to three passages of four days before considering them negative for FCV.
Genome extraction, capsid gene amplification and sequencingGenome extraction was performed with PureLink® Spin Column-Based Kit (Invitrogen® Life Technologies, USA). Reverse transcription was performed with Superscript III enzyme (Invitrogen®) according to the manufacturer's instructions. For conserved regions A-B of the capsid, a Nested-PCR was performed with primer pairs Cali1/Cali2 (position 5322–6246) and Cali3/Cali4 (position 5514–5991) as described by Marsilio et al.18 For variable regions C-F of the capsid, PCR reaction was initially conducted with primer pairs 8F/8R (position 6401–7080) described by Ohe et al.19 Only samples testing negative were subsequently attempted with the additional primers Percp1/Percp2 (position 6406–6913)20 and primer pair DEG-f – GGCCWGAYACMACMATHCC (position 6572–6592) and DEG-r – GCTYCYTCHCCAATKCCAGT (position 6922–6902), designed for this study.
The DEG-f/DEG-r PCR product was synthesized in 20μL of total solution containing the following: 5μL of cDNA, 1μL of both DEG-f and DEG/r 20pmol (Invitrogen®), 11.75μL of H2O DNAse/RNAse free (Gibco®), 2μL of dNTP (dATP, dTTP, dCTP and dGTP) 2.5mM (Invitrogen®), 2.5μL of 10× PCR Buffer (Invitrogen®), 1.5μL of MgCl2 50mM (Invitrogen®) and 0.25μL of Platinum Taq DNA Polymerase II (5U/μL) (Invitrogen®). Thermal-cycling conditions consisted of DNA denaturation at 94°C for 2min, followed by 35 cycles of denaturation at 94°C for 30s, primer annealing at 54°C for 45s, a primer extension at 72°C for 45s and a final extension at 72°C for 5min. Felocell CVR-C (Pfizer Animal Health, USA) was used as positive control. The position and sequence of the primers used for RT-PCR and nested RT-PCR amplifications were based on F9 prototype (GenBank access number: M86379). All the amplicons were purified using the QIAquick® PCR purification Kit (Qiagen® CA) and sequenced in both directions using the Big Dye Terminator® kit (Applied Biosystems CA).
In order to compare the sequences of this study with other published sequences, 104 FCV capsid sequences available in GenBank were selected. Among these prototypes, 13 Brazilian isolates from a molecular study performed in Southern Brazil were included.21 These were the only Brazilian FCV capsid sequences available so far in GenBank. Prototypes were aligned using the CLUSTAL W method into BioEdit® program22 and analyzed using DAMBE software23 for the exclusion of equal sequences.
Using PhyML 3.0 program,24 a Maximum Likelihood (ML) tree was reconstructed in an online web server.25 A heuristic tree search was performed using the SPR branch-swapping algorithm. To estimate the reliability of the obtained topology, we used the approximate likelihood-ratio test (aLRT)26 based on a Shimodaira-Hasegawa-like procedure. An aLTR value of 0.90 was considered significant. The trees were evaluated with Fig Tree v1.3.1 program.27
An amino acid (AA) sequence analysis between Brazilian isolates (from this study and from Southern Brazil) and F9 vaccine strain was performed with the MEGA 5.1 software program.28 An intragroup analysis calculating the divergence among isolates from both multicat long-life household and public short-term animal shelter also was performed with the MEGA 5.1.
Nucleotide sequences generated in this study were deposited in GenBank with the following accession numbers: KY427039 – KY427052 for the conserved region A-B and KY427032 – KY427038 and KY427053 for de variable region C-F.
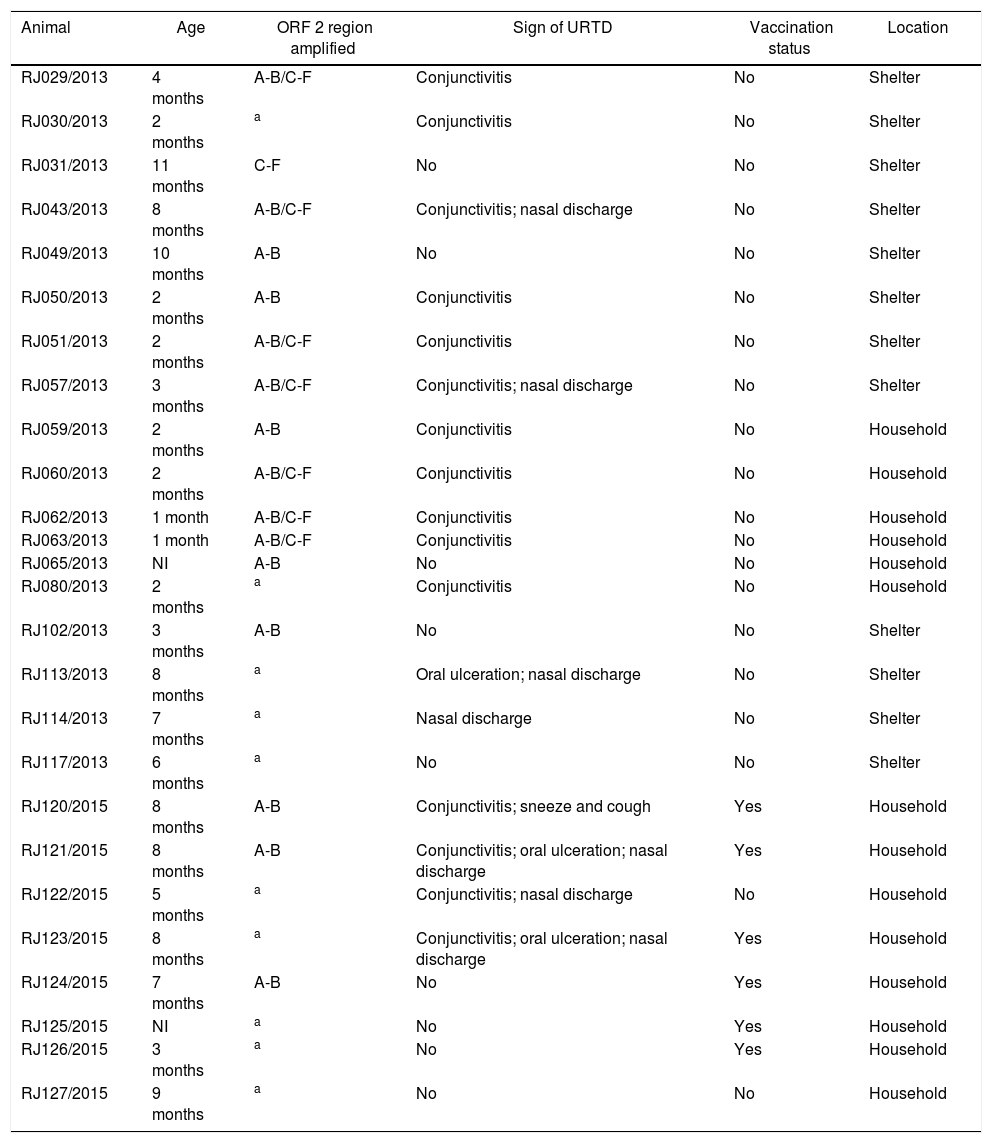
ResultsFourteen of the twenty-six samples were collected from multicat life-long households and twelve samples were collected from public short-term animal shelters. Six cats were vaccinated, however the vaccine schedule could not be taken. Cats’ age varied from one to eleven months-old. Regarding the clinical signs in the collection moment, fifteen cats had conjunctivitis, three had oral ulceration and eight had respiratory signs (sneeze and nasal discharge). Nine were asymptomatic, from which five were positive for FCV (Table 1).
Information from individual medical record and location from cats sampled in this study.
| Animal | Age | ORF 2 region amplified | Sign of URTD | Vaccination status | Location |
|---|---|---|---|---|---|
| RJ029/2013 | 4 months | A-B/C-F | Conjunctivitis | No | Shelter |
| RJ030/2013 | 2 months | a | Conjunctivitis | No | Shelter |
| RJ031/2013 | 11 months | C-F | No | No | Shelter |
| RJ043/2013 | 8 months | A-B/C-F | Conjunctivitis; nasal discharge | No | Shelter |
| RJ049/2013 | 10 months | A-B | No | No | Shelter |
| RJ050/2013 | 2 months | A-B | Conjunctivitis | No | Shelter |
| RJ051/2013 | 2 months | A-B/C-F | Conjunctivitis | No | Shelter |
| RJ057/2013 | 3 months | A-B/C-F | Conjunctivitis; nasal discharge | No | Shelter |
| RJ059/2013 | 2 months | A-B | Conjunctivitis | No | Household |
| RJ060/2013 | 2 months | A-B/C-F | Conjunctivitis | No | Household |
| RJ062/2013 | 1 month | A-B/C-F | Conjunctivitis | No | Household |
| RJ063/2013 | 1 month | A-B/C-F | Conjunctivitis | No | Household |
| RJ065/2013 | NI | A-B | No | No | Household |
| RJ080/2013 | 2 months | a | Conjunctivitis | No | Household |
| RJ102/2013 | 3 months | A-B | No | No | Shelter |
| RJ113/2013 | 8 months | a | Oral ulceration; nasal discharge | No | Shelter |
| RJ114/2013 | 7 months | a | Nasal discharge | No | Shelter |
| RJ117/2013 | 6 months | a | No | No | Shelter |
| RJ120/2015 | 8 months | A-B | Conjunctivitis; sneeze and cough | Yes | Household |
| RJ121/2015 | 8 months | A-B | Conjunctivitis; oral ulceration; nasal discharge | Yes | Household |
| RJ122/2015 | 5 months | a | Conjunctivitis; nasal discharge | No | Household |
| RJ123/2015 | 8 months | a | Conjunctivitis; oral ulceration; nasal discharge | Yes | Household |
| RJ124/2015 | 7 months | A-B | No | Yes | Household |
| RJ125/2015 | NI | a | No | Yes | Household |
| RJ126/2015 | 3 months | a | No | Yes | Household |
| RJ127/2015 | 9 months | a | No | No | Household |
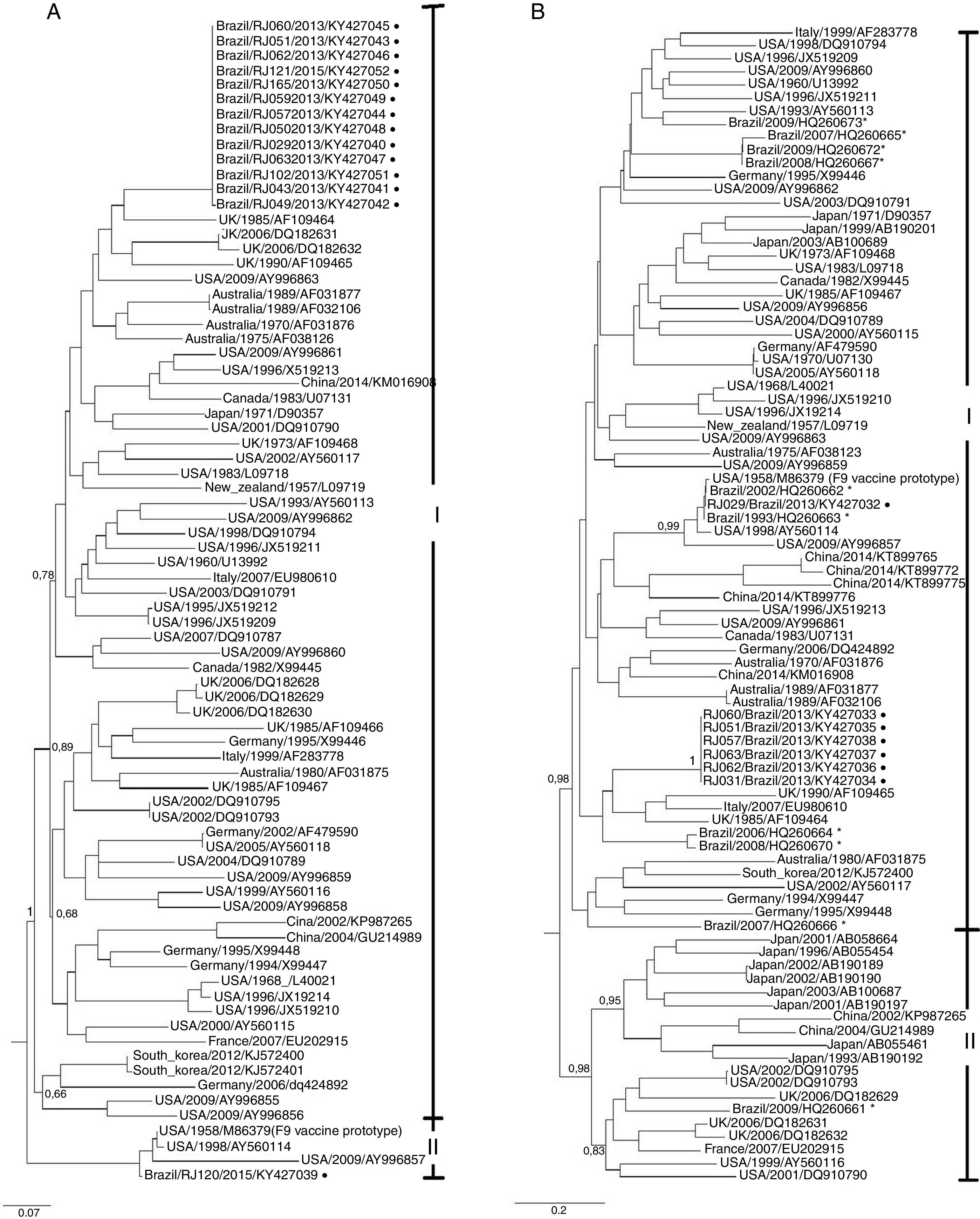
After all twenty-six samples passed for three passages in cell culture, eleven samples showed cytopathic effect. However, all the twenty-six isolates were submitted to genome extraction and then to RT-PCR for both conserved and variable region. Twenty-two isolates were amplified using the Cali1/Cali2 and Cali3/Cali4 primers and fourteen consensus sequences of the conserved regions A-B were of sufficient quality for phylogenetic analysis. Furthermore, eight isolates amplified with 8F/8R primers, twelve with Percp1/Percp2 primers and seven with DEG-f/DEG-r primer. Nevertheless, only eight isolates consensus sequences of the variable regions C-F were of sufficient quality and were used in phylogenetic analysis (Fig. 1).
ML trees of feline calicivirus isolated in this study along with most of the FCV prototypes available so far based on the ORF2 nucleotide sequences. (A) Phylogenetic tree of a 467bp partial fragment of the capsid conserved region A-B; (B) phylogenetic tree of a 529bp partial fragment of the capsid variable region C-F. Isolates from this study are represented with black circles and prototypes from Southern Brazil are represented with*.
Nucleotide identity calculated from isolates of this study and F9 vaccine prototype revealed an identity ranging from 73.4% to 96.1% on regions A-B and from 71.2% to 99% on region C-F. Samples RJ029/2013 and RJ120/2015 showed a high similarity to F9 prototype (99% and 96.1% respectively) and were considered F9-like sequences.11,29 Only sample RJ120/2015 was taken from a vaccinated cat, but interval between last vaccination and sample collection is not known. The divergence within groups based on regions C-F was 15.4% among isolates from public short-term animal shelters and 0.00% among isolates from multicat life-long households.
For the conserved regions A-B, phylogenetic analysis revealed two clusters (I and II) with an aLTR value of 1.00, as showed in Fig. 1. Most of the isolates from this study grouped in cluster I whilst sample RJ120/2015 grouped in cluster II with F9 vaccine prototype and other three prototypes form United States.
For the variable regions C-F, phylogenetic analysis also suggested two clusters (I and II) with an aLTR value of 0.98 (Fig. 1). Most of the Brazilian isolates were grouped in cluster I, except one which grouped in cluster II. Cluster II was also subdivided into two subclusters: one with most Chinese and Japanese isolates and a second with isolates from elsewhere in the world. Sample RJ029/2013 clustered with vaccine prototype F9, other two Brazilian isolates and three isolates from United States. The association between geographical distribution and/or clinical signs and distribution of isolates in different clusters was not observed.
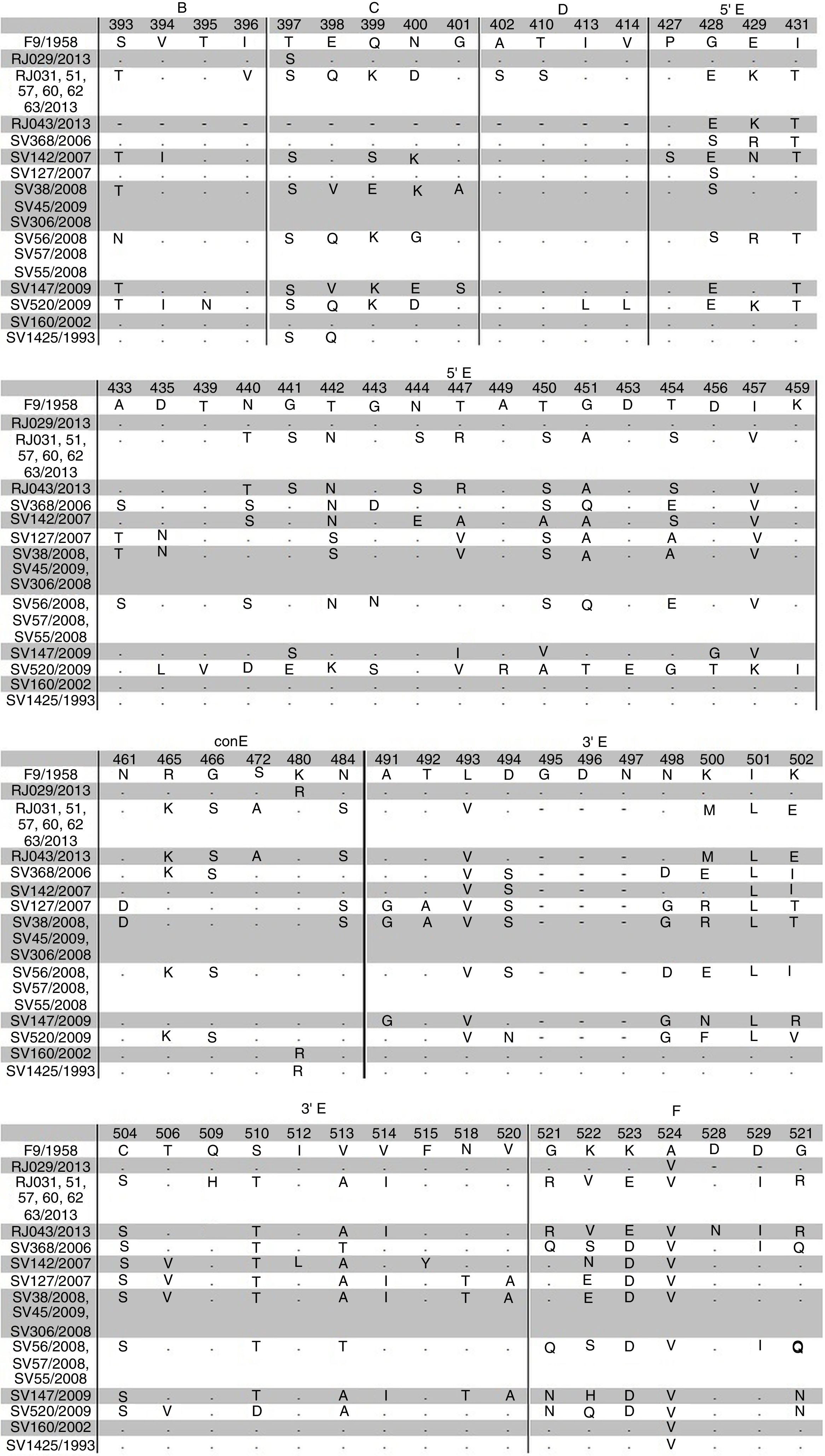
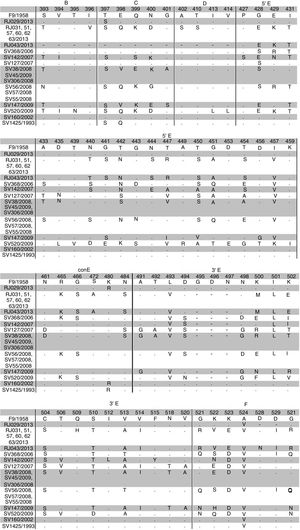
Alignment of the deduced amino acid sequences from regions B-F was performed and sequences from this study were compared with F9 prototype and other Brazilian strains. This analysis revealed a high variability in the regions C and E with AA changes in five residues in region C and 45 residues in region E (Fig. 2). Furthermore, D region is more conserved and ConE showed little variability with AA changes in five residues. The isolate RJ029/2013 and other two isolates of a previous Brazilian study (sv1425/1993, sv160/2002)21 had a close relationship with vaccine strain. They revealed AA substitution only at position 397, 398, 480 and 524.
Comparative amino acid sequence analysis of conserved B, D and part of F regions and variable C and E regions of FCV ORF2 among sequences from this study, other Brazilian isolates and F9 vaccine strain. Sequence numbering is based on Seal et al.7 Only the sites where sequences diverged were present in the figure. Isolates with equal amino acid sequence are in the same line. Sites where there are no sequences are indicated with a (-).
This is the first study using two different models for managing cats (multicat life-long household and public short-term animal shelter) and two different regions of capsid gene (conserved and variable regions) of FCV among cat populations in Brazil. For molecular characterization, the use of three distinct primer pairs to the capsid hypervariable region was necessary because the target sequence has high variability among FCV variants and the use of these primers combination allowed the characterization of 30% of the isolates. The same difficulty was reported by Coyne et al.,13 Hou et al.15 and Afonso et al.,30 that described a failure rate of 10–45% in generate quality FCV capsid PCR products, especially from most variable regions. The use of new-generation sequence strategies is recommended in order to increase the number of high quality sequences.31
Isolates from this study demonstrated a low level of viral diversity within multicat life-long households and a greater diversity level in public short-term animal shelters. This is in accordance to Radford et al.,32 Coyne et al.13 and Hou et al.15 that reported a high diversity of FCV isolates in shelters probably associated with multiple virus introductions arriving with cats from the community over time. However, a high diversity of variants was also reported in four endemically infected cat households33 and in a cross-sectional study in Europe.30 This high diversity of FCV is believed to be driven by a positive selection in the immunodominant 5′ and 3′ HVR of the capsid gene associated with a variable population immunity.30,33
FCV is considered to belong to a single antigenically diverse serotype34 and, so far, many studies have tried to classify the different variants based on hypervariable region E of the genome. Sato et al.35 and Sun et al.36 have proposed a classification dividing FCV isolates into two distinct genogroups based on phylogenetic analyses of capsid gene. Genogroup I include global isolates and genogroup II includes only isolates from Japan and China. Other study20 suggested that FCV forms two different clusters designated Major and Minor strains. However, most of the phylogenetical analysis typically results in a “star-like” phylogeny with little statistical support for grouping FCV strains in sub-species clusters either divide them into spatial, temporal or clinical grounds.15,21,37–39 The highest evolutionary rates of the virus lead to a saturation of evolutionary signals rapidly leading to a loss of phylogenetic resolution over relatively short time periods.38
The phylogenetic analysis of this study showed two clusters (I and II) with a significant aLTR value for the variable region and isolates from this study and the most of Brazilian isolated were close related with FCV genogroup I. As previously reported by,36 Chinese and Japanese isolates formed a single subgroup in genogroup II probably due the geographical proximity between these two countries.36
In the current study, an association between geographical distribution and/or clinical signs clustering in phylogenetic tree was not observed. This is in accordance to previous studies, where formation of a cluster corresponding to disease manifestation was not able to be noted neither for classical signs FCV strains nor for FCV-SVD strains.6,40,41
The 5′ and 3′ HVR and C region of Brazilian isolates demonstrated a high variability. The amino acids coded by 5′HVR are the target for neutralizing antibodies.10 Furthermore, isolates from this study showed the absence of three amino acids at 3′ HVR when compared with prototype F9. Henzel et al.21 also reported the absence of these amino acids in the isolates from Southern Brazil and suggested an evolution of the field isolates. FCV has limited ability for spread geographically and rare variants are transmitted over relatively large distances. Isolates may have evolved regionally with low dissemination capacity.38 Variants are usually confined to very close regions.15
RJ029/2013 and RJ120/2015 isolates were close related to the F9 vaccine strain, although RJ029/2013 was taken from an unvaccinated cat. Interval between last vaccination and sample collection from cat numbered as 120 was not known. The detection of vaccine virus post-vaccination is a rare event reported for two to five days in oral discharge.42 However, it is supposed that both isolates evolved from F9 vaccine strain. Vaccine-like viruses possibly exist in the field29 and previous studies have reported a rare occurrence of F9-like viruses among cats.32,38 These vaccine-derived viruses occasionally shed following vaccination appear at a low level in the cat population and seem to have a limited potential to persist in population.15,30
Sample RJ029/2013 was taken from a four months-old cat and RJ120/2015 was from an eight months-old cat and both cats had mild clinical conjunctivitis. Cat numbered as 120 also had respiratory signs. Both isolates also clustered with prototypes collected from cats with upper respiratory signs (USA/2009/AY996857 and USA/2009/AY996862) and eye lesion and upper respiratory signs (USA/1998/AY560114).43 Further, two Brazilian isolates (Brazil/1993/HQ260663 and Brazil/2002/HQ260662) clustered with RJ029/2013 and RJ120/2015. Clinical signs from these isolates were not informed.21 Because of considerable variability of FCV isolates, vaccine-induced immunity do not protect against field viruses,11 even if they belong to the same genogroup of the vaccine strain.19 This way, cats can become infected with field strains, although such cats are asymptomatic or have mild disease.12 In this study, one asymptomatic and vaccinated cat was positive for FCV. Outbreaks of FCV and occurrence of FCV-VSD are usually reported in vaccinated cats.5,38,44
ConclusionIn conclusion, the variants from this study belong to FCV genogroup I and the variants circulating in public short-term animal shelter demonstrated a high variability because of the relatively rapid turnover of carrier cats constantly introduced of multiple viruses into this location over time. Furthermore, F9-like virus can be isolated from unvaccinated cat with respiratory and ocular signs.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by the “Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro” (FAPERJ) (grant No. E-26/110.600/2014), by the “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior” (CAPES) and by the Plataforma de Sequenciamento de DNA of the Universidade Federal Fluminense.