Ovine/caprine ureaplasmas have not yet been assigned a species designation, but they have been classified into nine serotypes. Herein ureaplasmas were searched for in 120 samples of vulvo vaginal mucous from sheep and 98 samples from goats at 17 farms. In addition, semen samples were collected from 11 sheep and 23 goats. The recovered ureaplasma were from sheep and goats from animals without any reproductive disorder symptoms, but not all animals presented positive cultures. In sheep, 17 (68%) cultures of vulvovaginal mucous were positive for ureaplasma and 11 (27%) samples of semen presented positive cultures in animals with clinical signs of orchitis, balanoposthitis or low sperm motility. In goats four ureaplasma isolates were obtained from vulvovaginal mucus, but the semen samples were all negative. The isolates were submitted to Pulsed-field gel electrophoresis methodology and their 16S rRNA genes were sequenced. Fifty percent of ureaplasma recovered from sheep allowed for PFGE typing. Eleven isolates showed eight profiles genetically close to the bovine ureaplasmas. The 16S rRNA gene sequencing showed differences or similarities of isolates from sheep and goats, and the reference strains of bovine and human ureaplasma. Four clinical isolates from sheep were grouped separately. The studied ureaplasma isolates showed to be a diverse group of mollicutes.
The reduced genome of mollicutes codes about 700 proteins, and the few genes for DNA repair facilitate the genomic and antigenic variations. This helps to better understand their biological diversity and pathogenesis. Otherwise, the small genome facilitates the comparison of most species with the 16S rRNA gene sequences available in GenBank. The detection of genotypic variability of poorly studied mollicutes helps better organize their taxonomy and the inter-species and intra-species differences.1
Animal and human origin ureaplasma have the same basic metabolic features with about 750kpb in their genome, with a G-C% content of 26.9–28.0 in human isolates and 29.7–30.2 in bovine isolates. Both colonize the respiratory and urogenital tracts. Currently there are 2 species of human origin and five species isolated from bovine, canine, feline or avian hosts.2
Ovine/caprine ureaplasmas have not yet been assigned a species designation. Only the serological characteristics are known and they are classified in nine serotypes.3–5 Livingston6 and Ball7 studied ureaplasmas isolated from sheep in experimental inoculations. Serotype IX was characterized as causing infertility and abortion in sheep. However little is known about the molecular characteristics of these ureaplasma.8
In the present study, we used PFGE and 16S rRNA gene sequencing methodologies to determine genotypic differences among field isolates of ureaplasma recovered from reproductive tracts of male and female goats and sheep from Brazil.
Materials and methodsMicroorganismsUreaplasma diversum ATCC 49782 and ATCC 49783 (bovine ureaplasma) and one U. diversum isolate from bull semen were obtained from the Universidade de São Paulo (USP) collection. Mycoplasma ovine/caprine serogroup 11 AY121094; Ureaplasma canigenitalium D78648; Ureaplasma cati D78949; Ureaplasma gallorale U622937; U. diversum D78650; Ureaplasma felinum D78651; U. gallorale U62937; Ureaplasma urealyticum L40490; Ureaplasma serovar 5 U06097; Ureaplasma serovar 7 U06094 sequences deposited in the GeneBank were also used. The strains from USP collection were used for all genetic comparisons. The strains deposited in the GeneBank were used only to 16S rRNA sequencing comparisons.
Sample collectionOne hundred and twenty samples of vulvo vaginal mucous from ovine and 98 from caprine were obtained from 17 farms: 15 from the state of São Paulo, one from Pernambuco and one from Minas Gerais. The samples were collected from September 2005 to May 2007. Fifty percent of each group of animals was healthy and the remainder presented clinical signs, such as vaginal discharge, vaginitis, infertility, repeat breeding, abortions or stillbirths.
The vulva of each animal was previously cleaned with water and dried with a paper towel, and then sampled with a swab. The swab was directed dorsally before being redirected cranially, and then wiped over the cranial vaginal mucosa. In addition, semen samples were collected from 11 caprine (seven healthy) and 23 ovine (12 healthy). Animals with orchitis, testicular degeneration and low sperm motility were included. Semen samples were obtained by the electro-ejaculation method and transported to the laboratory at 4°C for culturing. Briefly, swab samples were homogenized in the broth, filtered through 0.45μm nitrocellulose membranes, and diluted to 10−4 in fresh ureaplasma broth. The Ureplasma agar plates were inoculated on the surface with a 200μL of each sample dilution. The cultures were incubated at 37°C in 5% CO2 up to 30 days.9
Pulsed-field gel electrophoresis (PFGE)The isolates and the ATCC strains were subcultured in 2mL ureaplasma medium at 37°C10 and expanded to 120mL of culture. Just after pH shifting of each broth, they were centrifuged for 75min at 16,300×g, then washed twice with PBS, followed by centrifugation for 25min at 20,600×g before being resuspended in 400μL of Tris-EDTA (10mM Tris–HCl, 1mM EDTA, pH 8.0). The DNA from the pelleted cells was extracted in agar blocks and digested with SalI, as described by Marois11 and Buzinhani.12 The PFGE was performed using 1.2% ultrapure agarose gel (Bio-Rad) in 0.5% TBE buffer (90mM Tris–borate, and 1mM EDTA) at 14°C in a CHEF Mapper DR-III (Bio-Rad). The program was standardized and carried out in three steps for PFGE of products digested with SalI enzyme. In the first step, the products were electrophoresed for 1h in 1% gel without and Lambda standard (Pharmacia) at an electrical pulse time (PT) of 1–4s for 8h. In the second step, gels were run for 10h at PT of 4–40s. Finally, the gels were stained with ethidium bromide (0.1μg/mL) for 60min and photodocumented with UV light. The size of the fragments was measured using the Vilber Lourmat photodocumentation program. Dendrograms were constructed using the BioNumerics program. The Unweighted Pair Group Method with Arithmetic Mean (UPGMA) was used to determine the genetic relationship between isolates and strains, applying the Dice similarity coefficient (%).
Sequencing of the 16S rRNA gene and phylogenetic analysesCultures of ureaplasma in 200mL of UB medium were used for DNA extraction according to the method described by Boom.13 The specific primer set F16S and R16 amplifies nearly the entire 16S rDNA sequence of mollicutes and was used as described by Harasawa and Cassell.14 PCR products (10μL) were then electrophoresed in 1% agarose gel with 10μg/mL of ethidium bromide and visualized with UV light (Vilber Lourmat, Germany). After confirmation of the targeted product fragment, PCR products were purified with GFX PCR DNA and Gel Band Purification Kit (Amersham) and quantified using a Low Mass DNA Ladder (Invitrogen). Purified PCR products were sequenced according to the MegaBACE 1000 protocol, using the DYEnamic ET Dye Terminator kit (with Thermo Sequenase II DNA Polymerase). Sequences were analyzed in the Sequence Analyzer software, Base Caller Cimarron 3.12. The 16S rRNA gene sequences were compared to sequences in the GenBank databases using the BLASTn algorithm (Mycoplasma ovine/caprine serogroup 11 AY121094; Ureaplasma canigenitalium D78648; Ureaplasma cati D78949; Ureaplasma gallorale U622937; Ureaplasma diversum D78650; Ureaplasma felinum D78651; Ureaplasma gallorale U62937; Ureaplasma urealyticum L40490; Ureaplasma serovar 5 U06097; Ureaplasma serovar 7 U06094). Phylogenetic analyses were performed using a BioNumerics program. Briefly, a multiple sequence alignment of the DNA sequences was constructed using the ClustalW.15 A phylogenetic tree was obtained using the neighbor-joining method with Tajima-Nei correction. The data set was resampled 1000 times to generate bootstrap values.
ResultsCulturesThe recovered ureaplasma were from sheep and goats from animals without any symptom of a reproductive disorder, but not all animals presented positive cultures. In sheep 17 (68%) cultures of vulvovaginal mucous were positive for ureaplasma. In semen, 11 (27%) samples also presented ureaplasmal growth of animals with clinical signs of orchitis, balanoposthitis or low-sperm motility. Samples of vulvovaginal mucus of goats presented four positive cultures for ureaplasma. However the cultures from all 23-goat semen samples were negative for ureaplasma.
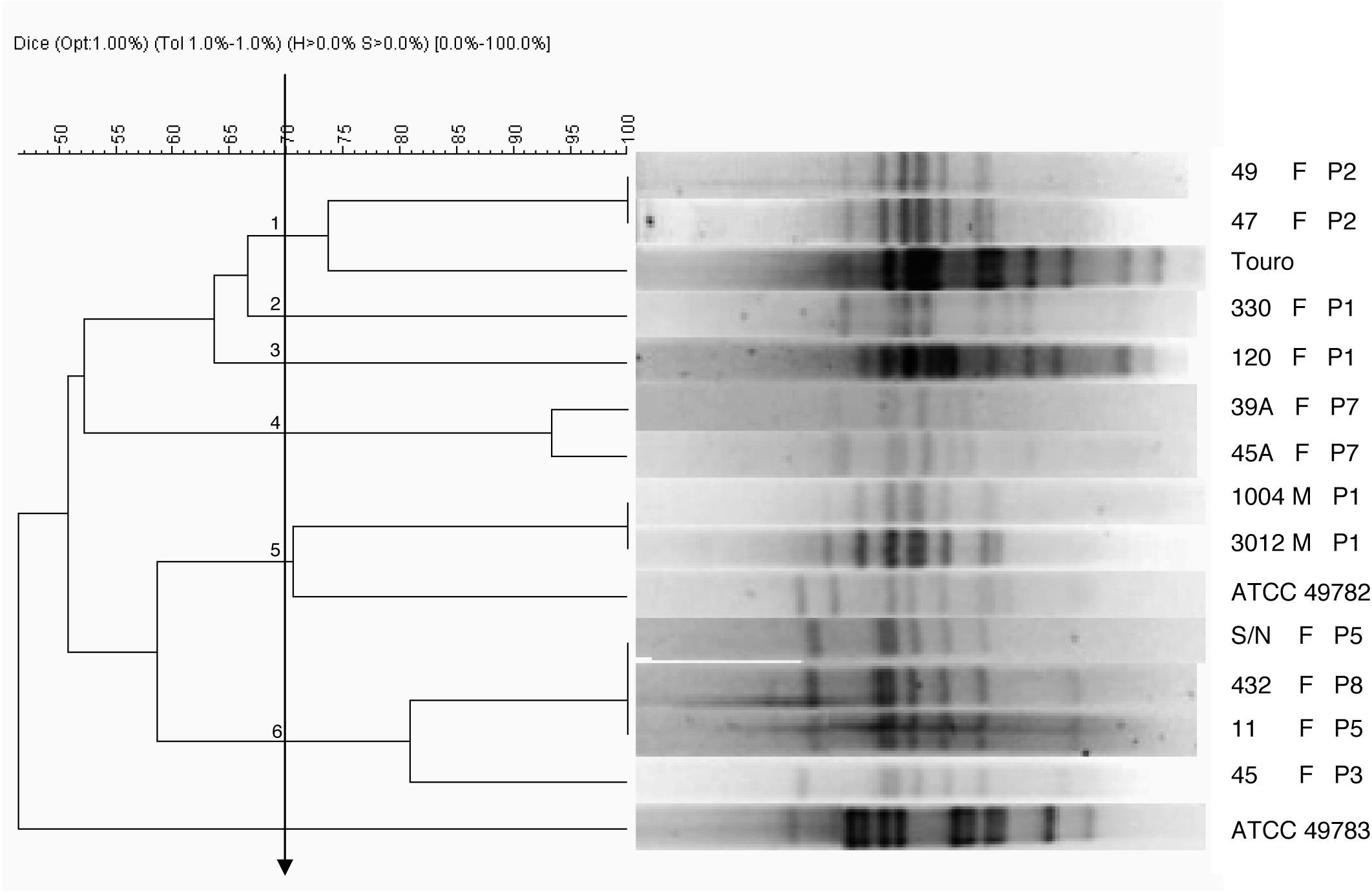
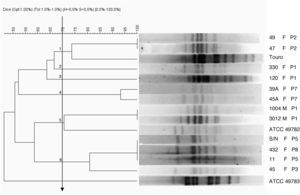
Pulse field gel electrophoresis (PFGE)Only 24 isolates were used for PFGE assay because some lost viability. The 24 clinical isolates of ureaplasma and reference strains were subjected to PFGE. However, 12 (50%) of ovine origin ureaplasma showed fragments for analysis (10 from vulvovaginal mucous and two from semen). The dendrogram obtained by DICE coefficient from the SalI enzyme profiles obtained using a cutoff ≥70% similarity allowed for grouping the clinical isolates into six clusters with similarities ranging from 74 to 100% (Fig. 1). Despite being the same species, the bovine ureaplasma were previously isolated and used as a control, and the ATCC strains were grouped in different clusters. This bovine ureaplasma and U. diversum ATCC 49782 were grouped in different clusters. However, the isolates from sheep and U. diversum ATCC 49783 were grouped separately from other studied ureaplasmas (clinical isolates of ureaplasma from sheep, bovine ureaplasma obtained from bull and the reference strain ATCC 49782).
Dendrogram obtained by UPGMA cluster analysis of DNA fragments from ureaplasmas isolated from sheep. Ureaplasma bovine isolate (Touro 1) and Ureaplasma diversum reference strains (ATCC 49782 and ATCC 49783) were also digested by the enzyme SalI. F, female; M, male; P, farm. Arrow: cutoff “Cluster”: 1, 2, 3, 4, 5 and 6.
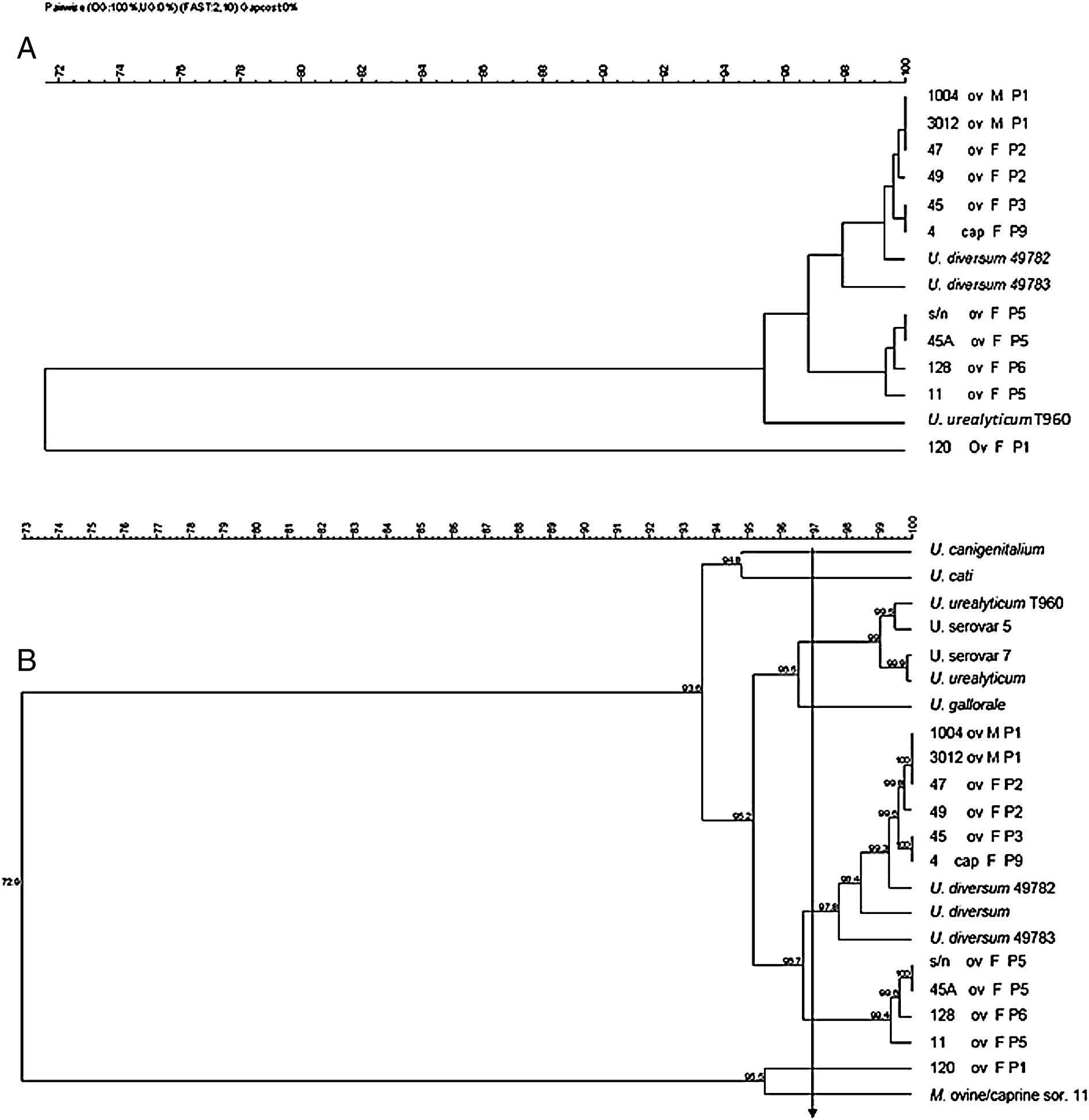
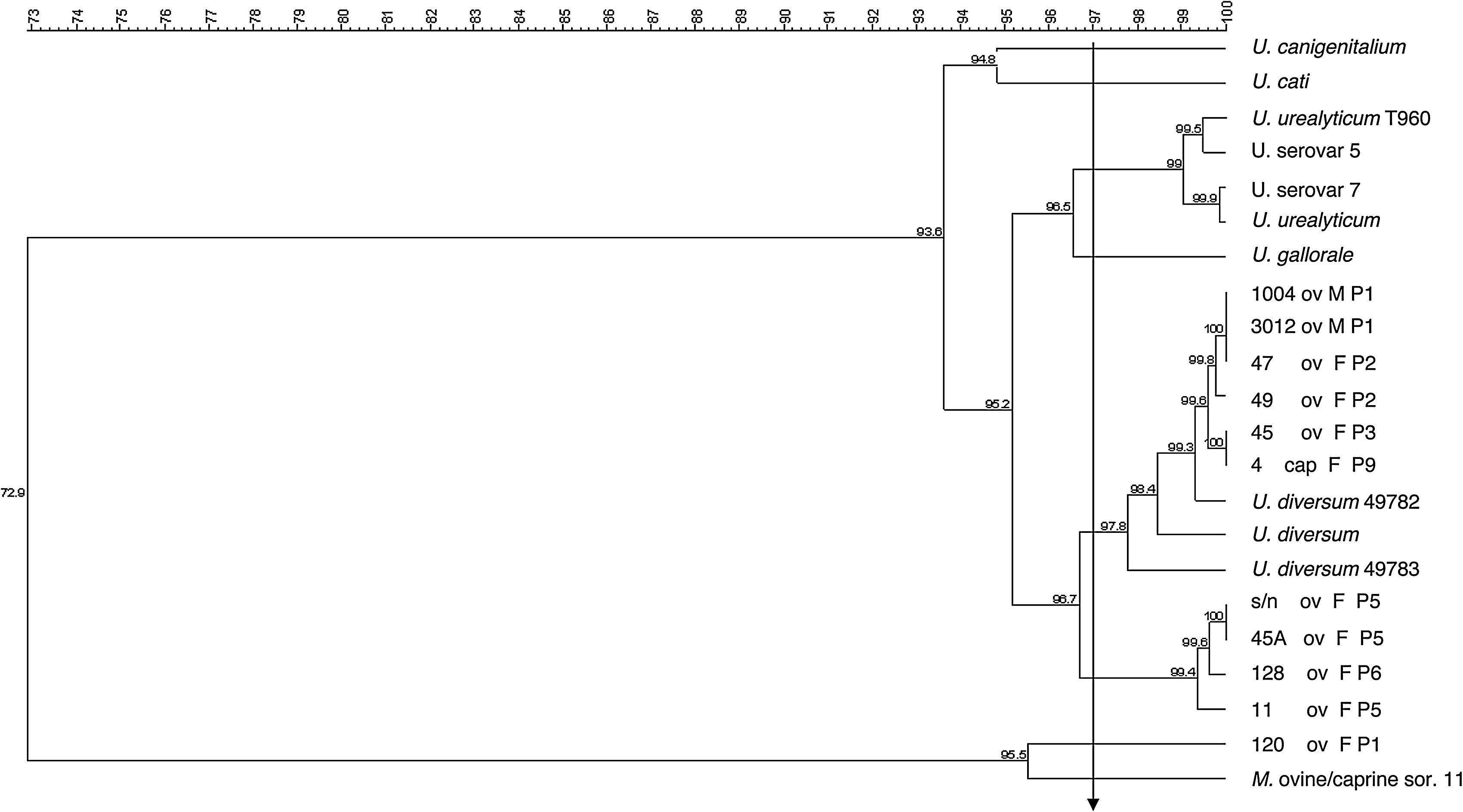
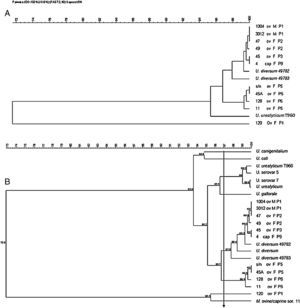
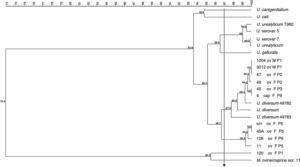
The dendrogram in Fig. 2 was obtained from the 16S rRNA gene sequencing of isolates and reference strains with a cutoff ≥97% similarity, also showing separation of genera Ureaplasma and Mycoplasma. The ureaplasma group was divided into cluster A with the human ureaplasma T960, and cluster B with ureaplasmas of large and small ruminants. In sub-cluster C, six clinical isolates were grouped with U. diversum (ATCC 49782 and 49783), with 98–100% homology. The sub-cluster D grouped four isolates of sheep (s/n, 45, 128 and 11), not related to U. diversum (Fig. 2). The dendrogram in Fig. 3 shows human and other animal ureaplasma including species recovered from goats and sheep grouped with U. diversum. However, the same four isolates (s/n, 45, 128 and 11) were in another group of other ureaplasmas. Mycoplasma isolated from sheep obtained in the present study (unpublished data) was grouped together with Mycoplasma ovine/caprine serogroup 11 (Fig. 3).
Dendrogram obtained from sequences of 16S rRNA gene of ureaplasma isolates from goats and sheep. Ureaplasma bovine isolate (120), Ureaplasma urealyticum and U. diversum reference strains (ATCC 49782 and ATCC 49783) were also included. Arrow: cutoff ≥97% (suitable for intraspecific similarity analysis). ov, sheep; cap, goat; F, female; M, male; P, farm.
Dendrogram obtained from the sequences of 16S rRNA gene of isolates of ureaplasma isolates from goats and sheep and ureaplasmas isolated from humans and animals were included for comparison. Ureaplasma bovine isolate (120) and Mycoplasma ovine/caprine serogroup 11 were also included. Arrow: cutoff ≥97% (suitable for intraspecific similarity analysis). ov, sheep; cap, goat; F, female; M, male; P, farm.
Mycoplasmas are among infectious agents found in the urogenital tract of sheep and goats.16 M. agalactiae, M. serovar 11, M. arginini M. capricolum, M. mycoides LC, A. laidlawii and Ureaplasma spp were recovered from the genital tract of goats and sheep. These mollicutes have been proposed to be potential infectious agents in genital infections of mentioned hosts. In Brazil these data are practically unknown.17 Since the first report of urogenital disease in sheep associated with ureaplasma,18 a few similar studies have been published.19,20 Mycoplasmosis has been more thoroughly studied in cattle than in sheep and goats; however, pathogenicity due to mollicutes in sheep and goats has been better associated.
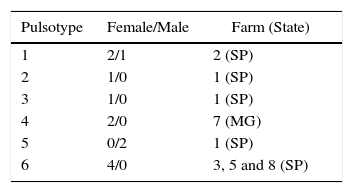
In the present study, 50% of recovered ureaplasma from sheep were genetically typified using PFGE. Marois11 and Buzinhani12 reported that 100% of the isolates of Mycoplasma synoviae and U. diversum, respectively, did not allow for typing with PFGE methodology. Buzinhani12 mentioned the absence of fragments due to the low DNA content. In fact, ureaplasma grows fast, but is sensitive to pH shift and is fragile to osmotic shock due to the absence of the cell wall. These growth features make it difficult to obtain enough ureaplasmal DNA. Nevertheless, herein, the PFGE of studied ureaplasma allowed for obtaining eight different profiles and showed that the typified isolates were genetically close to bovine ureaplasmas. Isolates from farm 1 were grouped among different pulsotypes in the present analysis (Table 1). It is noteworthy that a correlation between pulsotype of the isolates and animal health could not be a related, as well as an association of the isolates and virulence factors. Isolates obtained from farm 3, 5 and 8 were clustered in same pulsotype. This can be explained by the geographical proximity of the farms. Due to the proximity of the farms, there may be an exchange of animals.
The 16S rRNA gene was sequenced to detect the divergence of ureaplasma isolates from sheep, goats, bovine and humans. This allowed for obtaining phylogenetic trees and grouping the microorganisms. Our findings are consistent with other studies that show a similarity between ureaplasmas isolated from these animals.3,5 However, four of the clinical (s/n, 45, 128 and 11) isolates of sheep were grouped in a separate group. However, four isolates of sheep were not grouped with the ureaplasma species, which could be indicative of a new species. Although the isolates were grouped as U. diversum, it is not yet possible to conclude that they are the same species.
More studies are needed to better characterize the genetic differences and similarity reported here. The techniques used in this study can be applied in monitoring and screening of herds. The viability of these methods helps in monitoring these microorganisms and reducing economic loss in goat and sheep ranching. The diagnosis of mycoplasma in the reproductive tract of sheep and goats is widely practiced, making it difficult to control these microorganisms.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (grant 06/56855-0). We thank Aricelma P. França for valuable technical assistance, as well as AcademicEnglishSolutions.com for revising the English.