Although the consumption of fresh and minimally processed vegetables is considered healthy, outbreaks related to the contamination of these products are frequently reported. Among the food-borne pathogens that contaminate vegetables is Listeria monocytogenes, a ubiquitous organism that exhibits the ability to survive and multiply at refrigerated temperatures. This study aimed to evaluate the occurrence of L. monocytogenes in vegetables as well as the antimicrobial resistance of isolates. The results showed that 3.03% of samples were contaminated with L. monocytogenes, comprising 2.22% of raw vegetables and 5.56% of ready-to-eat vegetables. Multiplex PCR confirmed the virulence potential of the isolates. Antimicrobial resistance profiling showed that 50% of the isolates were susceptible to the antibiotics used. The resistance of one isolate to penicillin G, a commonly employed therapeutic agent, and the presence of serotype 4b, a serotype commonly associated with food-borne outbreaks, could be potential health hazards for consumers.
Food safety is an increasingly relevant issue in the daily lives of consumers. The search for a healthier diet and, at the same time, faster preparation has favored the consumption of fresh and ready-to-eat vegetables. Because they are not subjected to treatments that considerably reduce microbiological hazards, these essential foods are potential vehicles for the transmission of pathogenic microorganisms.1
To inhibit microbial multiplication and ensure adequate conservation, certain vegetables are stored and transported at cool temperatures. However, these conditions facilitate the growth of some microbial pathogens, such as Listeria monocytogenes, a psychotropic microorganism that is highly relevant to public health.1,2
L. monocytogenes is a ubiquitous bacterium that can be found in the irrigation water, soil and fertilizer used on farms and in decaying plant matter, making the presence of this bacterium in vegetables a continual risk.
The disease caused by L. monocytogenes, known as listeriosis, is particularly troublesome for vulnerable populations. This group of people, including pregnant women and their fetuses, the very young and the elderly, is particularly susceptible to invasive listeriosis, with mortality rates ranging between 20 and 40%.3
Outbreaks and sporadic cases of listeriosis have been associated with the contamination of various food items, including milk; soft cheese; meat and meat products; vegetables; seafood products; ready-to-eat foods4; and, recently, cantaloupes.5
It is well established that food product contamination is associated with food-processing environments harboring L. monocytogenes and subsequent post-processing transfer to finished products.6,7
In Brazil, food-borne listeriosis outbreaks have not been documented, but in recent studies, the presence of L. monocytogenes has been described in several products, including ready-to-eat vegetables, such as watercress and escarole8; leafy salads9; chopped kale and a mixture of spring onion/parsley10; salad mix, lettuce, collard greens, a mix for yakisoba, watercress, escarole, cabbage, spinach, and a mix for sukiyaki.11
L. monocytogenes is naturally susceptible to a range of antibiotics that act on Gram-positive bacteria.12 Human strains of L. monocytogenes are sensitive to a group of antibiotics that includes penicillin, ampicillin, amoxicillin, gentamicin, erythromycin, tetracycline, rifampicin, co-trimoxazole, vancomycin and imipenem.13,14 However, most strains of L. monocytogenes show natural resistance to current fluoroquinolones and cephalosporins, and especially those of the third and fourth generations, such as cefotaxime and cefepime, and to fosfomycin, oxacillin and lincosamides.13
Clinicians usually treat listeriosis with aminopenicillins in combination with an aminoglycoside, such as gentamicin. Additionally, in cases in which reduced sensitivity or resistance to beta-lactams is encountered, a number of agents that are active against Gram-positive bacteria may be used.14
Studies performed by the Clinical and Laboratory Standards Institute (CLSI)15 using human strains and, to a lesser extent, foodstuff strains have not revealed an increase in resistance to antibiotics among the circulating strains of L. monocytogenes.16,17 However, since the isolation of the first multi-resistant strain of L. monocytogenes in 1988, interspecies variation in antimicrobial susceptibilities has been reported among Listeria species.12,18
The purpose of this study was to verify the occurrence of L. monocytogenes in raw, frozen and ready-to-eat vegetables commercialized in different locales in Salvador, BA, Brazil, and to characterize L. monocytogenes strain using the multiplex PCR method. Furthermore, the resistance/susceptibility of the L. monocytogenes strains to eight antibiotics used in human and veterinary medicine was investigated.
Materials and methodsSample collectionRaw, frozen and ready-to-eat vegetables were purchased at a local supermarket and at fast food outlets in Salvador, BA, in northeastern Brazil, from October 2013 through January 2014. Overall, 132 samples were collected, comprising 45 raw vegetables, 33 frozen vegetables and 54 ready-to-eat vegetables (salads). The raw vegetables included lettuce, broccoli, white cabbage, purple cabbage and arugula (nine samples of each). The frozen vegetables consisted of mixed vegetables (peas, carrots, green beans and maize), peas and broccoli (11 samples of each). The ready-to-eat vegetables included salads containing carrot, purple cabbage and lettuce; salads containing lettuce, beet and purple cabbage; and salads containing purple cabbage, white cabbage, lettuce and beets (18 samples each). Representative sampling was ensured by taking samples from the most consumed brands that contained at least one of the most consumed vegetables in Brazil.19
Detection of L. monocytogenesFor the isolation of L. monocytogenes, approximately 25g of each sample was homogenized with 225mL of half-concentrated Fraser broth (FB; Merck, Darmstadt, Germany) in a stomacher (240bpm; ITR model 1204, series 126; São Paulo, SP, Brazil) for 2min in a class II biosafety cabinet (Labconco Purifier Class IIb, Total Exhaust, model 36210-04, certified ISO 9002; Labconco Corporation, Kansas City, MO, USA). This homogenate was then incubated at 30°C for 24h. An aliquot of 1mL was transferred to tubes containing FB supplemented with Fraser selective supplement and incubated at 37°C for 48h. The cultures were streaked onto plates containing the Listeria agar Ottaviani & Agosti (ALOA™, Laborclin, Pinhais, PR, Brazil) and incubated at 37°C for 24h.20 Afterward, 3–5 suspect colonies (blue, diameter less than 3mm and with a regular white halo) were selected for confirmation. The confirmation of L. monocytogenes colonies in isolation media was based on several methods, including Gram staining and measurement of hemolytic activity on sheep blood agar (Columbia agar supplemented with 5% defibrinated horse blood; HiMedia, São Paulo, SP, Brazil), the carbohydrate utilization pattern (0.5% mannitol, 0.5% rhamnose and 0.5% xylose), the catalase reaction and tumbling motility.21 One L. monocytogenes Scott A (serotype 4b; ATCC 15313) positive control and one uninoculated-medium negative control were used for each set of concurrently analyzed samples.
Antigenic characterization was performed in the Laboratory of Bacterial Zoonoses (LABZOO/IOC/FIOCRUZ) using somatic and flagellar serotyping antisera produced in the same laboratory, as described by Seeliger and Höhne.22 All strains are deposited in the Collection of Listeria (CLIST) from LABZOO and maintained in BHI broth with 20% glycerol at −20°C.
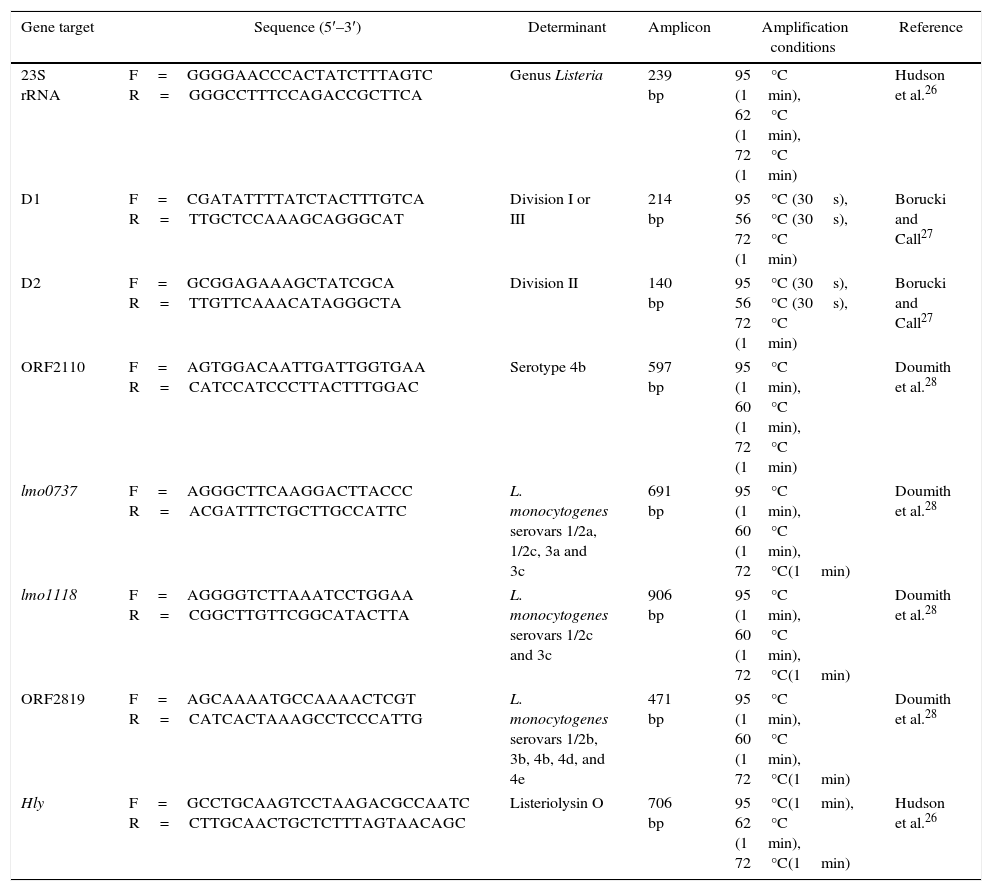
Multiplex PCR confirmation of isolatesMultiplex PCR was performed to confirm genus, species, lineage and serotypes, and the conditions and the set of primers are summarized in Table 1. The DNA was extracted using the DNeasy Blood & Tissue kit® (Qiagen, Hilden, Germany) following the manufacturer's instructions. The PCR assays in a final volume of 50μL were done using the following: 1× reaction buffer, MgCl2 1.5mM, 0.4mM of each dNTP, 10pmol/μL of each primer and 0.5U/μL of HotStar®Taq polymerase (Qiagen, Hilden, Germany). Standard strains of L. monocytogenes ATCC 19115 (serotype 4b), ATCC 19111 (serotype 1/2a), ATCC 19112 (serotype 1/2c) and CDC F4976 (serotype 1/2b), were used as positive controls, and Listeria innocua ATCC 12612 was used as a negative control.
Primers, amplicon size and amplification conditions for the PCR assays.
| Gene target | Sequence (5′–3′) | Determinant | Amplicon | Amplification conditions | Reference |
|---|---|---|---|---|---|
| 23S rRNA | F=GGGGAACCCACTATCTTTAGTC R=GGGCCTTTCCAGACCGCTTCA | Genus Listeria | 239 bp | 95°C (1min), 62°C (1min), 72°C (1min) | Hudson et al.26 |
| D1 | F=CGATATTTTATCTACTTTGTCA R=TTGCTCCAAAGCAGGGCAT | Division I or III | 214 bp | 95°C (30s), 56°C (30s), 72°C (1min) | Borucki and Call27 |
| D2 | F=GCGGAGAAAGCTATCGCA R=TTGTTCAAACATAGGGCTA | Division II | 140 bp | 95°C (30s), 56°C (30s), 72°C (1min) | Borucki and Call27 |
| ORF2110 | F=AGTGGACAATTGATTGGTGAA R=CATCCATCCCTTACTTTGGAC | Serotype 4b | 597 bp | 95°C (1min), 60°C (1min), 72°C (1min) | Doumith et al.28 |
| lmo0737 | F=AGGGCTTCAAGGACTTACCC R=ACGATTTCTGCTTGCCATTC | L. monocytogenes serovars 1/2a, 1/2c, 3a and 3c | 691 bp | 95°C (1min), 60°C (1min), 72°C(1min) | Doumith et al.28 |
| lmo1118 | F=AGGGGTCTTAAATCCTGGAA R=CGGCTTGTTCGGCATACTTA | L. monocytogenes serovars 1/2c and 3c | 906 bp | 95°C (1min), 60°C (1min), 72°C(1min) | Doumith et al.28 |
| ORF2819 | F=AGCAAAATGCCAAAACTCGT R=CATCACTAAAGCCTCCCATTG | L. monocytogenes serovars 1/2b, 3b, 4b, 4d, and 4e | 471 bp | 95°C (1min), 60°C (1min), 72°C(1min) | Doumith et al.28 |
| Hly | F=GCCTGCAAGTCCTAAGACGCCAATC R=CTTGCAACTGCTCTTTAGTAACAGC | Listeriolysin O | 706 bp | 95°C(1min), 62°C (1min), 72°C(1min) | Hudson et al.26 |
The amplified fragments were subjected to electrophoresis on a 2% agarose gel (in TBE buffer), stained with ethidium bromide solution (10mg/mL) (Sigma, São Paulo, Brazil) and visualized with a UV transilluminator coupled with a digital gel imaging system (Kodak EDAS 290).
Antimicrobial resistance of the isolatesAntimicrobial resistance was assessed by a disk diffusion assay according to CLSI guidelines15 using the breakpoints of Staphylococcus species resistance because no resistance criteria exist for Listeria susceptibility testing in the CLSI guidelines.23–25
Cells were grown at 35°C for 24h in tryptic soy broth (TSB; HiMedia, São Paulo, SP, Brazil), suspended in a saline solution and diluted to 0.5 points on the McFarland scale (ca. 108cfu). The suspension of cells was inoculated in Mueller-Hinton (MH) agar (HiMedia, São Paulo, SP, Brazil) using a swab. Bacterial growth was recorded after 24h of incubation at 35°C. One Staphylococcus aureus (ATCC 33591) positive control was used for each set of analyzed samples. The isolates were tested against a panel of eight antimicrobial agents: penicillin G (PEN) (10IU), erythromycin (ERY) (15μg), tetracycline (TET) (30μg), oxacillin (OXA) (1μg), cefoxitin (CEF) (30μg), vancomycin (VCM) (30μg), streptomycin (STR) (10μg) and ciprofloxacin (CIP) (5μg). The antibiotic discs were purchased from Laborclin (Pinhais, PR, Brazil). The zones of inhibition were measured (mm) at 24h.
Results and discussionPrevalence of L. monocytogenes in vegetablesIn this work, we detected and identified L. monocytogenes in raw and ready-to-eat vegetables purchased at supermarkets located in the city of Salvador, Brazil, to show the existing risk to consumers.
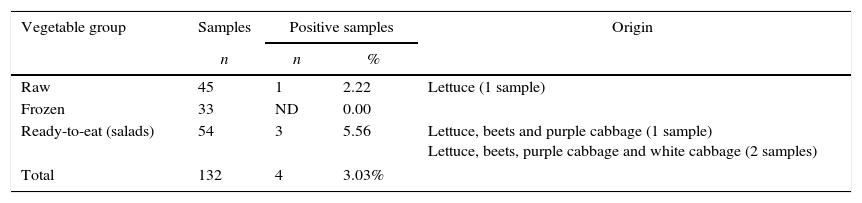
Of the 132 samples analyzed, L. monocytogenes was isolated from four (3.03%) samples, of which one (2.22%) came from raw vegetables and three (5.56%) came from ready-to-eat vegetables (salads) (Table 2).
Occurrence of L. monocytogenes in raw, frozen and ready-to-eat vegetables.
| Vegetable group | Samples | Positive samples | Origin | |
|---|---|---|---|---|
| n | n | % | ||
| Raw | 45 | 1 | 2.22 | Lettuce (1 sample) |
| Frozen | 33 | ND | 0.00 | |
| Ready-to-eat (salads) | 54 | 3 | 5.56 | Lettuce, beets and purple cabbage (1 sample) Lettuce, beets, purple cabbage and white cabbage (2 samples) |
| Total | 132 | 4 | 3.03% | |
ND, not detected.
Lettuce was the only raw vegetable contaminated by L. monocytogenes, whereas two types of mixed vegetable salads presented the microorganism. No sample of frozen vegetables was contaminated by L. monocytogenes.
The four obtained isolates recovered from the raw and ready-to-eat vegetable samples were identified as L. monocytogenes serotype 4b by both the conventional antigenic characterization and molecular serotyping methodologies. Multiplex PCR targeting the listeriolysin O genes (hly) of the L. monocytogenes recovered from the vegetable samples confirmed the virulence potential of the isolates.
The presence of L. monocytogenes in many types of raw and ready-to-eat vegetables intended for human consumption has been clearly demonstrated in many countries. In Brazil, the incidence of L. monocytogenes in vegetables has been reported in many studies by a number of researchers. The results found in the present work demonstrated higher levels of detection of L. monocytogenes in salads than the levels previously reported by Froder et al. (0.55%),9 Oliveira et al. (3.7%),10 and Sant’ana et al. (3.1%).11
In countries other than Brazil, various results have also been reported for the occurrence of L. monocytogenes in vegetables. In Santiago, Chile, the bacterium was isolated from 10.2% of salad samples29; in Malaysia, from 22.5% of salad samples30; in Spain, from 4.18% of vegetable samples31; in Marmara, Turkey, from 13.6% of fresh vegetable samples32 and in Germany, from only four samples of 1001 investigated vegetable samples.33
Work performed by Moreno et al.31 in Valencia, Spain, showed that nine lettuce samples were contaminated by L. monocytogenes, with counts between 2.85 and 3.55log10 viable cells/g of food. Additionally, only one positive sample, found in a dish called a ‘Four Seasons Salad,’ yielded a high number of viable cells, with a value of 4.35log10 cells/g of food.
The presence of L. monocytogenes in vegetables, verified in the present study and in other studies reported by various authors, is cause for concern, as listeriosis cases are increasing at the global level, in many cases due to the cross-contamination of processed foods.31 Therefore, Good Agricultural Practices and Good Manufacturing Practices must be addressed to guarantee the safety of food for consumers.
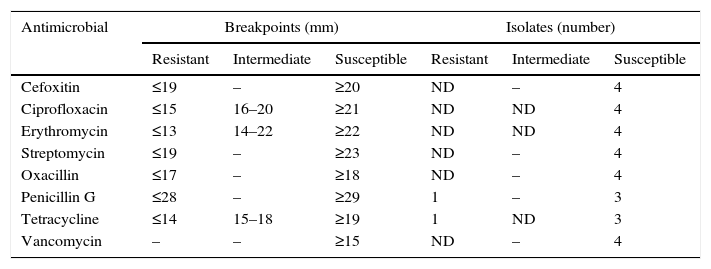
Antimicrobial resistanceIn the current study, all L. monocytogenes isolates were susceptible to erythromycin (ERY), oxacillin (OXA), cefoxitin (CEF), vancomycin (VCM), streptomycin (STR) and ciprofloxacin (CIP) (Table 3).
Resistance/susceptibility of L. monocytogenes isolated from vegetables.
| Antimicrobial | Breakpoints (mm) | Isolates (number) | ||||
|---|---|---|---|---|---|---|
| Resistant | Intermediate | Susceptible | Resistant | Intermediate | Susceptible | |
| Cefoxitin | ≤19 | – | ≥20 | ND | – | 4 |
| Ciprofloxacin | ≤15 | 16–20 | ≥21 | ND | ND | 4 |
| Erythromycin | ≤13 | 14–22 | ≥22 | ND | ND | 4 |
| Streptomycin | ≤19 | – | ≥23 | ND | – | 4 |
| Oxacillin | ≤17 | – | ≥18 | ND | – | 4 |
| Penicillin G | ≤28 | – | ≥29 | 1 | – | 3 |
| Tetracycline | ≤14 | 15–18 | ≥19 | 1 | ND | 3 |
| Vancomycin | – | – | ≥15 | ND | – | 4 |
ND, not detected.
Two L. monocytogenes isolates from ready-to-eat vegetables exhibited resistance to penicillin G (PEN) and tetracycline (TET).
In contrast, Kovacevic et al.34 found that L. monocytogenes recovered from ready-to-eat fish possessed reduced susceptibility to ciprofloxacin and two of the three clonal 1/2b L. monocytogenes isolates were resistant to streptomycin. However, like the results of present study, the authors observed no resistance to erythromycin.
Studies performed by Davis and Jackson25 with strains of L. monocytogenes recovered from diverse origins (i.e. clinical, animal, food, and environmental) showed similar results to those in the present work with regard to CIP and TET resistance. Also, Troxler et al.35 in Germany tested 87 strains of Listeria spp. (19 L. monocytogenes from human and sheep and avian origin), predominantly isolated in the USA and different European countries, and grouped L. monocytogenes and L. welshimeri as naturally sensitive to CIP.
However, according to Kovacevic et al.34, the extent of comparison between studies is hampered by differences in the methods used to verify resistance.
In general, antibiotic therapy is required for the treatment of listeriosis infections, with clinicians commonly using ampicillin in combination with gentamycin or using trimethoprim-sulfamethoxazole.36 In the present study, resistance to penicillin G, a commonly employed therapeutic agent, was observed for one isolate of L. monocytogenes.
According to Kovacevic et al.,34 only three isolates of L. monocytogenes recovered from 4668 clinical samples in France were resistant to streptomycin (STR). Two isolates were resistant to lower concentrations (4 and 6mg/mL), whereas one exhibited resistance at 256mg/mL.17 Regarding food and environmental strains, none of the 49 L. monocytogenes isolates from food and environmental samples tested in the U.S. in 2009 showed resistance to STR (1mg/mL). In 2010, another study performed in U.S. with L. monocytogenes isolated from catfish filets and their respective processing environments reported reduced susceptibility to STR (10mg) in 2% (2/80) of the isolates).23 Has been reported that sublethal exposure to triclosan, a broad-spectrum biocide used into a variety of commercial products, promotes resistance to various aminoglycosides, including gentamycin, kanamycin, streptomycin and tobramycin.34 In addition, resistance to low and high concentrations of the antibiotic has been suggested to be associated with possible ribosomal mutations and/or the production of 6-N-streptomycin adenylyltransferase, encoded by the aad6 gene.17,34 These finds is a cause for concern, considering that clinicians typically use aminopenicillins (e.g., ampicillin or amoxicillin) in combination with an aminoglycoside, such as gentamicin, for the treatment of invasive infections.13,36
Similar to the results presented in our study, Gómez et al.13 found that one strain (0.5%) of L. monocytogenes recovered from a stainless steel surface in Spanish industry was resistant to tetracycline and eight strains (3.9%) recovered from meat products showed reduced susceptibility to penicillin G. According to the authors, the effectiveness of tetracycline diminished in the last decades owing to the widespread existence of resistance genes, probably because of the extensive and prolonged use of these antimicrobials in human medicine and as growth promoters in animals. In addition, regarding fluoroquinolones, intermediate susceptibility to ciprofloxacin was found in the mentioned work for one strain of the pathogenic species. The authors concluded that all Listeria strains were highly sensitive to the preferred antibiotics used to treat listeriosis, although special mention was made of the reduced susceptibility of eight strains of L. monocytogenes (3.9%) and of seven strains of L. innocua (5.4%) to penicillin G.
The isolates of L. monocytogenes investigated in the present study did not show multi-resistance.
Work performed by Korsak et al.37 on 471 samples from different types of food and food-related sources in Poland demonstrated that one L. monocytogenes strain, isolated in 2005 from iced green beans, was resistant to tetracycline (MIC 16μg/ml). Chen et al.,23 who investigated ten samples of foods commercialized in China, reported similar results, with 2.7% of the isolates resistant to tetracycline and 8.1% resistant to penicillin. These results show that antimicrobial resistance in L. monocytogenes occurs at a low prevalence.
ConclusionThe presence of L. monocytogenes in fresh and ready-to-eat vegetables and the resistance of isolates to penicillin G, a commonly employed therapeutic agent, as verified in the present study, are cause for concern, as listeriosis cases are increasing at the global level. In addition, the presence of serotype 4b, more commonly associated with outbreaks, could be a potential health hazard for consumers.
Conflicts of interestThe content of this report solely reflects the opinions of the authors, and we report no conflicts of interest.
This research was supported in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). We wish to thank students Rodrigo de Castro Lisboa and Vanessa de Souza Rodrigues, from the Laboratório de Zoonoses Bacterianas (LABZOO/IOC/FIOCRUZ), Rio de Janeiro, RJ, Brazil, for help in PCR assay.