This work aimed to characterize 20 isolates obtained from upland rice plants, based on phenotypic (morphology, enzymatic activity, inorganic phosphate solubilization, carbon source use, antagonism), genotypic assays (16S rRNA sequencing) and plant growth promotion. Results showed a great morphological, metabolic and genetic variability among bacterial isolates. All isolates showed positive activity for catalase and protease enzymes and, 90% of the isolates showed positive activity for amylase, catalase and, nitrogenase. All isolates were able to metabolize sucrose and malic acid in contrast with mannitol, which was metabolized only by one isolate. For the other carbon sources, we observed a great variability in its use by the isolates. Most isolates showed antibiosis against Rhizoctonia solani (75%) and Sclerotinia sclerotiorum (55%) and, 50% of them showed antibiosis against both pathogens. Six isolates showed simultaneous ability of antibiosis, inorganic phosphate solubilization and protease activity. Based on phylogenetic analysis of the 16S rRNA gene all the isolates belong to Bacillus genus. Under greenhouse conditions, two isolates (S4 and S22) improved to about 24%, 25%, 30% and 31% the Total N, leaf area, shoot dry weight and root dry weight, respectively, of rice plants, indicating that they should be tested for this ability under field conditions.
Beneficial endophytic microorganisms can live inside plants without causing damage to the host. Among them, many bacteria colonize the intercellular spaces throughout the whole plant, including seeds.1 Intercellular spaces contain sources of carbohydrates and minerals, such as: nitrogen, phosphorus, calcium, potassium, and chlorine, as well as other metabolites as organic acids, facilitating bacterial development and colonizing.2
Among several microorganisms colonizing plant tissues, plant growth-promoting rhizobacteria (PGPR) were also found.3 These PGPRs provide beneficial effects, such as: mineral nutrition improvement, increase on the tolerance of biotic and abiotic stresses, root development promotion, and suppression of soil borne diseases.2
Also, these bacteria can be involved on nitrogen fixation, inorganic phosphate solubilization, iron complexation, phytohormones synthesis and plant pathogen control, involving many enzymes on these processes.2,4 Thus, plants under influence of PGPR can be greater, stronger, more productive and healthier.3,5,6 Besides, the assessment of biochemical characteristics in bacteria allows inferences about their adaptive capacity to the environment and about the plant–bacteria interaction, since it involves several genes and mechanisms, such as chemoattraction and biofilm formation, among other activities.3
Association between PGPRs and root of rice plants has been studied and many isolates showing effects on rice growth were found.5 The application of this technology on rice chain production can provide benefits for crop growth and reduce production costs, as well as helping on the reduction of environmental risks.5,7
Furthermore, the identification and characterization of bacteria as PGPR provide insights for understanding the composition of bacterial communities associated with rice plants grown under Cerrado conditions. However, studies with focus on the isolation and characterization of bacteria from Cerrado soils able to promote plant growth of upland rice are still scarce as well as, necessary for this crop. Thus, this study aimed to characterize bacterial isolates obtained from rice roots based on biochemical and genetic characteristics and, to determine their ability to promote plant growth aiming their use as inoculant for upland rice.
Material and methodsBacterial isolatesThe isolates evaluated in this study were obtained from rice roots by Rezende8 and are available at the Collection of Microorganisms and Multifunctional Fungi of Embrapa Rice and Beans.
Morphological characterization and Gram coloring assaysMorphological characterization was performed according to Vermelho et al.9 based on shape, border, surface, consistence and elevation of the colonies. Gram coloring was performed according to Louvet et al.10
Biochemical assaysAll biochemical trials were performed on a completely randomized design in triplicate under laboratory conditions. For the evaluation of carbon sources use (maleic acid, malic acid, nicotinic acid, d-arabinose, d-fructose, d-glucose, mannitol, mio-inositol, sucrose and sorbitol), each isolate was inoculated in Petri dishes containing solid King B medium added with each different carbon sources and incubated (28°C; 2 d). After incubation, we checked out the growth of the isolates according to Hungria et al.11
The activity of the enzymes citrate lyase and urease was measured through qualitative assays by inoculating the isolates on solid Simmons citrate and urea medium and observing the blue and red colors of the culture medium, respectively.9 Nitrate reductase and nitrogenase enzymes were qualitatively assayed in semi-solid medium.12 Catalase activity was assayed according to Vermelho et al.9 For amylase and protease activities, we observed the formation of a translucid halo around the colonies, formed by the isolates inoculated on M9 solid medium containing 1% of starch and 2% of milk protein, respectively.13 Cellulase activity was determined according to Cattelan et al.14
Solubilization of inorganic phosphate assay was performed using three replicates. Solubilization ability was confirmed based on the observation of a translucid halo around the colonies formed by the isolates inoculated on NBRI-P15 and Pikovskaya medium.16
Antagonistic testThe antagonistic ability of the isolates was performed using three replicates, under in vitro conditions against the fungi Sclerotinia sclerotiorum and Rhizoctonia solani on Petri dishes containing BDA medium according to Hammami et al.17
Clustering analysis of the isolates based on morpho-physiological dataMorphological, enzymatic activity, carbon source use and antibiosis data were transformed into a binary matrix and submitted to cluster analysis performed by the software NTSYSpc®23 using UPGMA as grouping algorithm and Jaccard as similarity coefficient.
16S rRNA gene sequencing analysisBased on the clustering analysis of the isolates based on morpho-physiological data 11 isolates (S2, S4, S6A, S17, S22, S26, S29, S37, S41, S63 and S105) were selected for 16S rRNA gene sequencing. The DNA of the isolates was obtained according to Laranjo et al.18 The 16S rRNA gene was amplified by PCR using the primers Y1 and Y3.18,19 The amplicons were purified and used on the sequencing reaction according to Laranjo et al.18 Sequences coding for the partial 16S rRNA gene of the isolates S2, S4, S6A, S17, S22, S26, S29, S37, S41, S63 and S105 was obtained and when submitted to the GenBank database (www.ncbi.nlm.nih.gov) received the accession numbers KX855959.1, KX855958.1, KX197211.1, KX197210.1, KX855960.1, KX197209.1, KX855957.1, KX197208.1, KX197214.1, KX197206.1 and KX197207.1, respectively.
Evaluation of plant growth promotionBased on genotypic characterization, four isolates were selected to be evaluated under greenhouse conditions as compared to a no-inoculated treatment (NI). Seeds of rice cv. Aimoré were sown in 8L pots filled with soil and arranged in a random block design with four replicates. At sowing time, seeds were inoculated with peat-based inoculant containing 1×108cellg−1 of each isolate and reference strain. Rice plants were harvested at 40 days after emergence (DAE) to determine leaf area (LA), root volume (RV), root dry weight (RDW), shoot dry weight (SDW) according to Ferreira20,21 and, the total N (N-Total) using the Kjedahl method, as described by Silva and Queiroz.22
Data analysisFor the 16S rRNA-based phylogeny study, the sequences obtained were submitted to NCBI BLAST against a non-redundant nucleotide database for getting homologous sequences.24 Sequences showing degree of similarity were aligned using the CLUSTAL W program.25 The evolutionary history was inferred by using the maximum likelihood method,26 with tree consensus being inferred from 500 replicates using bootstrap.27 All positions containing gaps and missing data were eliminated. There were a total of 1312 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.28
Data obtained from the greenhouse experiment were subjected to analysis of variance. When F was significant, the Scott–Knott test of means was applied with a 5% probability using SISVAR statistical software.
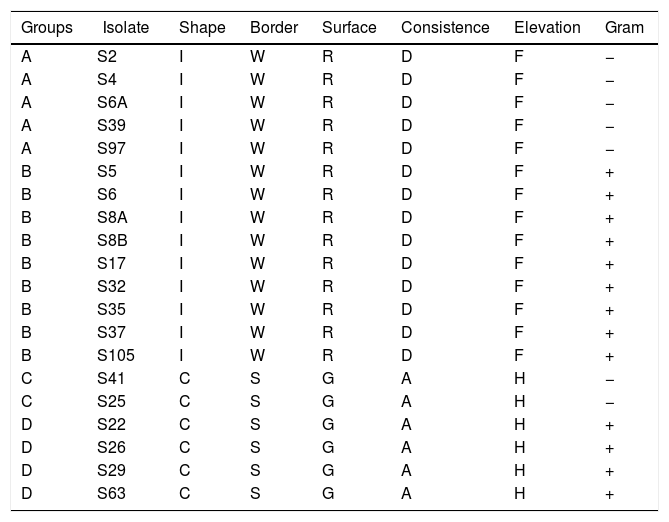
ResultsAnalysis based on the morphological characteristics of the isolates resulted in four different groups (A, B, C and D). The groups were composed by 5, 9, 2 and 4 isolates, respectively (Table 1). Isolates of groups A and B show the same morphological characteristics, except for Gram coloration, by which the isolates of group A are Gram−, while isolates of group B are Gram+. Similarly, isolates of the group C are Gram−, and isolates of group D are Gram+. Additionally, isolates of groups A and B show different morphological characteristics compared to isolates of groups C and D (Table 1).
Morphological characteristics on King B medium of endophytic isolates obtained from rice plants.
| Groups | Isolate | Shape | Border | Surface | Consistence | Elevation | Gram |
|---|---|---|---|---|---|---|---|
| A | S2 | I | W | R | D | F | − |
| A | S4 | I | W | R | D | F | − |
| A | S6A | I | W | R | D | F | − |
| A | S39 | I | W | R | D | F | − |
| A | S97 | I | W | R | D | F | − |
| B | S5 | I | W | R | D | F | + |
| B | S6 | I | W | R | D | F | + |
| B | S8A | I | W | R | D | F | + |
| B | S8B | I | W | R | D | F | + |
| B | S17 | I | W | R | D | F | + |
| B | S32 | I | W | R | D | F | + |
| B | S35 | I | W | R | D | F | + |
| B | S37 | I | W | R | D | F | + |
| B | S105 | I | W | R | D | F | + |
| C | S41 | C | S | G | A | H | − |
| C | S25 | C | S | G | A | H | − |
| D | S22 | C | S | G | A | H | + |
| D | S26 | C | S | G | A | H | + |
| D | S29 | C | S | G | A | H | + |
| D | S63 | C | S | G | A | H | + |
I, irregular; C, circular; W, wrinkled; S, smooth; R, rough; G, gentle; D, dry; A, aqueous; F, flat, H, high.
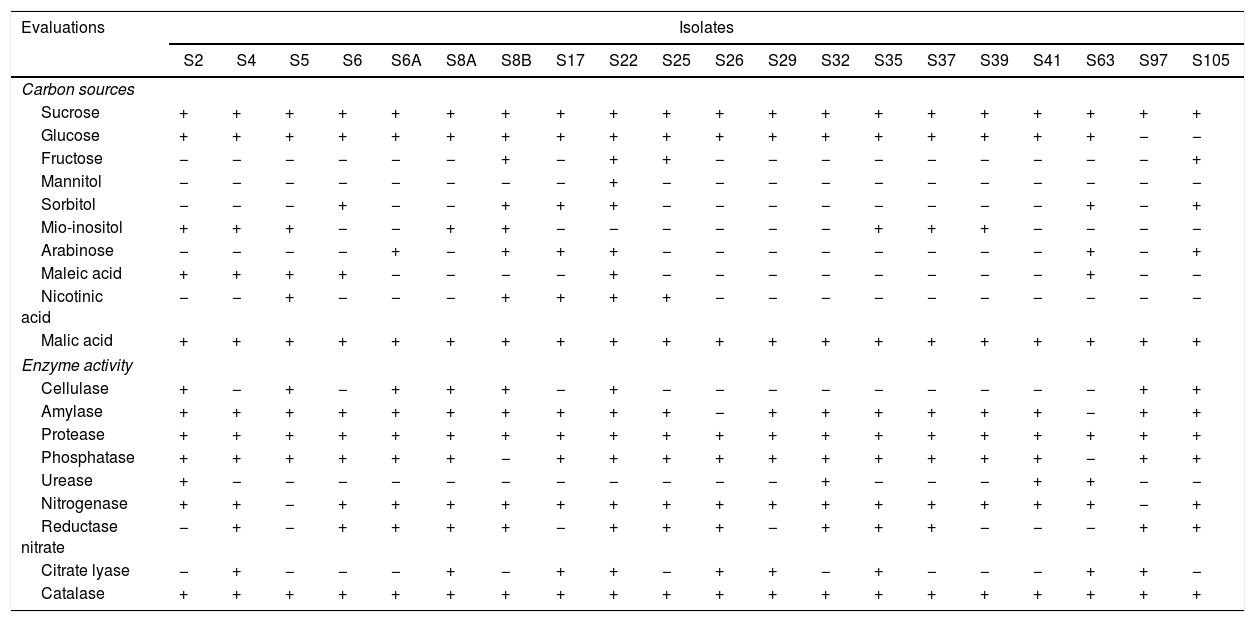
The results of carbon source use and enzyme activity showed a great metabolic diversity among the isolates (Table 2).
Metabolic characteristics of endophytic isolates obtained from rice plants.
| Evaluations | Isolates | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S2 | S4 | S5 | S6 | S6A | S8A | S8B | S17 | S22 | S25 | S26 | S29 | S32 | S35 | S37 | S39 | S41 | S63 | S97 | S105 | |
| Carbon sources | ||||||||||||||||||||
| Sucrose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − |
| Fructose | − | − | − | − | − | − | + | − | + | + | − | − | − | − | − | − | − | − | − | + |
| Mannitol | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| Sorbitol | − | − | − | + | − | − | + | + | + | − | − | − | − | − | − | − | − | + | − | + |
| Mio-inositol | + | + | + | − | − | + | + | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Arabinose | − | − | − | − | + | − | + | + | + | − | − | − | − | − | − | − | − | + | − | + |
| Maleic acid | + | + | + | + | − | − | − | − | + | − | − | − | − | − | − | − | − | + | − | − |
| Nicotinic acid | − | − | + | − | − | − | + | + | + | + | − | − | − | − | − | − | − | − | − | − |
| Malic acid | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Enzyme activity | ||||||||||||||||||||
| Cellulase | + | − | + | − | + | + | + | − | + | − | − | − | − | − | − | − | − | − | + | + |
| Amylase | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | − | + | + |
| Protease | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Phosphatase | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | − | + | + |
| Urease | + | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | + | − | − |
| Nitrogenase | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + |
| Reductase nitrate | − | + | − | + | + | + | + | − | + | + | + | − | + | + | + | − | − | − | + | + |
| Citrate lyase | − | + | − | − | − | + | − | + | + | − | + | + | − | + | − | − | − | + | + | − |
| Catalase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
All the isolates were able to use sucrose and malic acid as carbon source, while only the isolate S22 was able to use mannitol. Also, 45% and 35% of the isolates were able to use fructose and mio-inositol, respectively. Besides, the isolate S22 was able to use nine of the ten carbon sources evaluated, while the isolate S97 used just two of these sources (Table 2).
All the isolates showed catalase activity, while 90% of the isolates showed amylase, phosphatase and nitrogenase activity. The isolates S8A, S22, S35 and S105 showed activity of seven enzymes, and isolates S5 and S19 showed activity of four (Table 2).
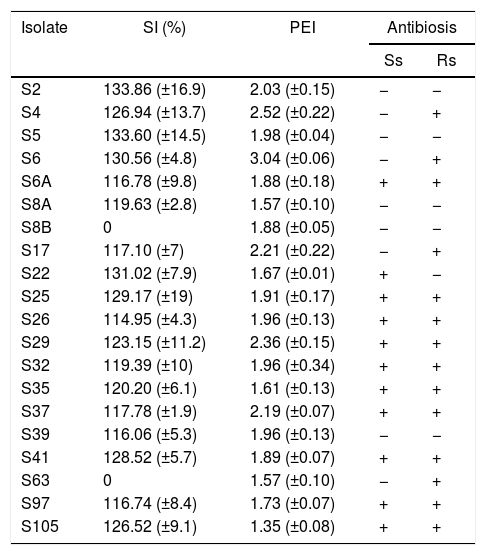
For the phosphate solubilization assay, only the results obtained from the Pikovskaya medium are shown, since we did not observe growth of the isolates in the NBRI-P medium. About 75% of the isolates were able to solubilize inorganic phosphate (Table 3). These isolates showed a SI of about 111% in average, with the greatest values been observed for isolates S2 (133.86%) and S5 (133.6%). Isolates S8b and S63 did not present solubilization ability.
Solubilization index (SI) for inorganic phosphate assayed in Pikovskaya medium, protease enzymatic index (PEI) and antibiosis assays against Sclerotinia sclerotiorum (Ss) and Rhizoctonia solani (Rs) of endophytic isolates obtained from rice plants. Mean values of three replicates.
| Isolate | SI (%) | PEI | Antibiosis | |
|---|---|---|---|---|
| Ss | Rs | |||
| S2 | 133.86 (±16.9) | 2.03 (±0.15) | − | − |
| S4 | 126.94 (±13.7) | 2.52 (±0.22) | − | + |
| S5 | 133.60 (±14.5) | 1.98 (±0.04) | − | − |
| S6 | 130.56 (±4.8) | 3.04 (±0.06) | − | + |
| S6A | 116.78 (±9.8) | 1.88 (±0.18) | + | + |
| S8A | 119.63 (±2.8) | 1.57 (±0.10) | − | − |
| S8B | 0 | 1.88 (±0.05) | − | − |
| S17 | 117.10 (±7) | 2.21 (±0.22) | − | + |
| S22 | 131.02 (±7.9) | 1.67 (±0.01) | + | − |
| S25 | 129.17 (±19) | 1.91 (±0.17) | + | + |
| S26 | 114.95 (±4.3) | 1.96 (±0.13) | + | + |
| S29 | 123.15 (±11.2) | 2.36 (±0.15) | + | + |
| S32 | 119.39 (±10) | 1.96 (±0.34) | + | + |
| S35 | 120.20 (±6.1) | 1.61 (±0.13) | + | + |
| S37 | 117.78 (±1.9) | 2.19 (±0.07) | + | + |
| S39 | 116.06 (±5.3) | 1.96 (±0.13) | − | − |
| S41 | 128.52 (±5.7) | 1.89 (±0.07) | + | + |
| S63 | 0 | 1.57 (±0.10) | − | + |
| S97 | 116.74 (±8.4) | 1.73 (±0.07) | + | + |
| S105 | 126.52 (±9.1) | 1.35 (±0.08) | + | + |
The general mean observed for PEI was about 1.92. Six isolates (S6, S4, S29, S17, S37 and S2) showed PEI greater than 2, while the isolate S105 showed the smallest value for PEI (Table 3).
On the antibiosis assays against S. sclerotiorum, 11 isolates were able to inhibit the growth of the fungi, while against R. solani 14 isolates inhibited the fungi growth. Besides, the isolates S6A, S25, S26, S29, S32, S35, S37, S41, S97 and S105 showed antibiosis against both fungi (Table 3).
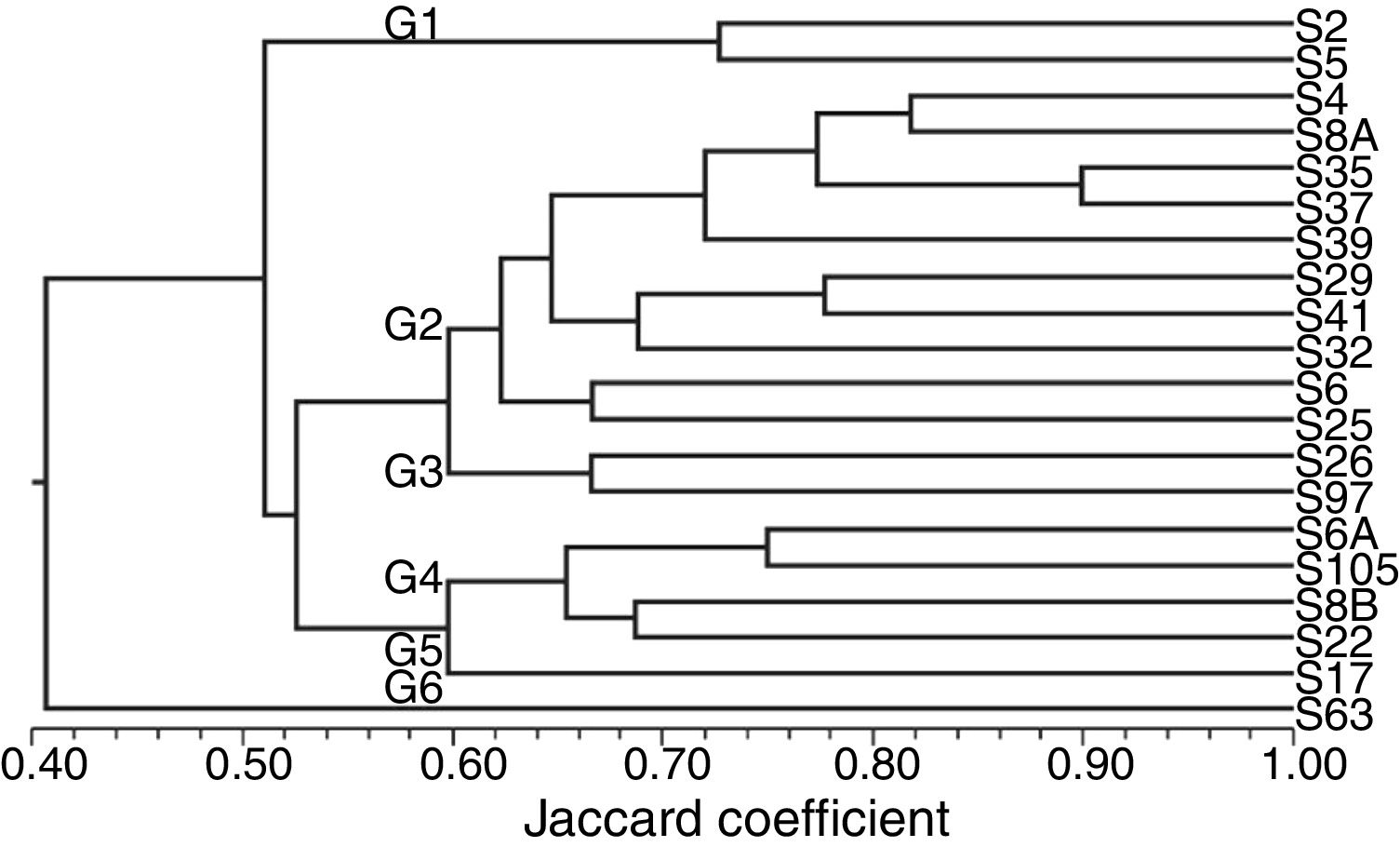
The similarity analysis based on morphological characteristics, enzymatic activity, carbon source use and antibiosis assay revealed six clusters formation at 60% of similarity, indicating great diversity among the 20 isolates regarding these traits (Fig. 1).
Consensus dendrogram obtained by combining the morphological, enzymatic activity, carbon source use and antibiosis data among 20 endophytic isolates obtained from upland rice plants. Dendrogram was generated by the algorithm UPGMA and similarity matrix obtained from the use of Jaccard coefficient.
The isolate S63, on group G6, showed a 40% similarity to the other isolates, followed by the isolates of the group G1. The greatest similarity was about 90%, observed between the isolates S5 and S37. These isolates grouped with other eight isolates on group G2 (Fig. 1).
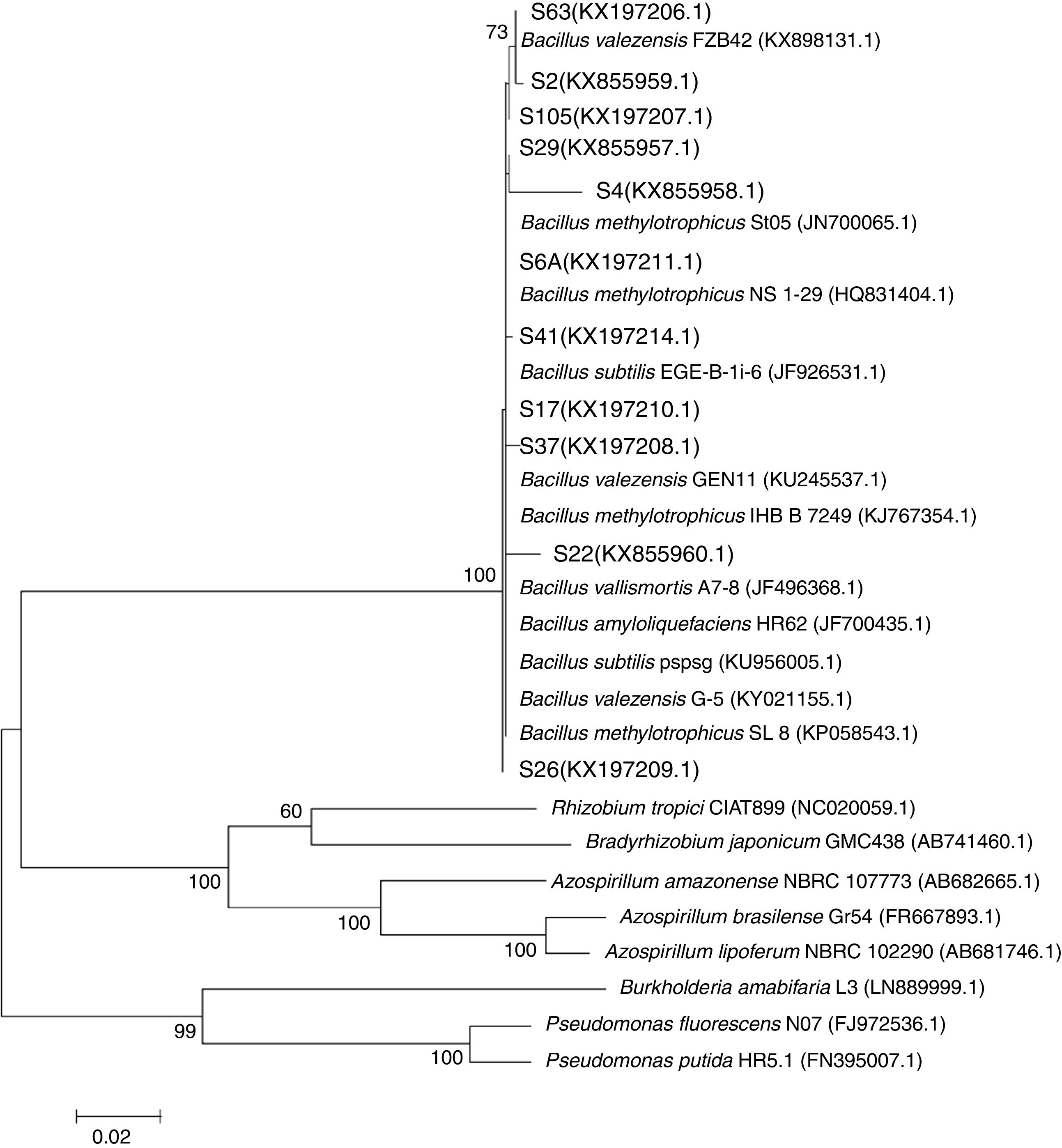
The 16S rRNA gene sequencing analysis revealed that all the evaluated isolates showed high similarity with the genus Bacillus as compared with other plant growth-promoting genera (Fig. 2).
Maximum likelihood phylogeny of the 16S rRNA gene showing the relationships among endophytic isolates obtained from rice plants (in bold) with different genera of plant growth-promoting rhizobacteria. GenBank accession numbers are shown in parentheses. Bar, 20nt substitutions per 1000nt.
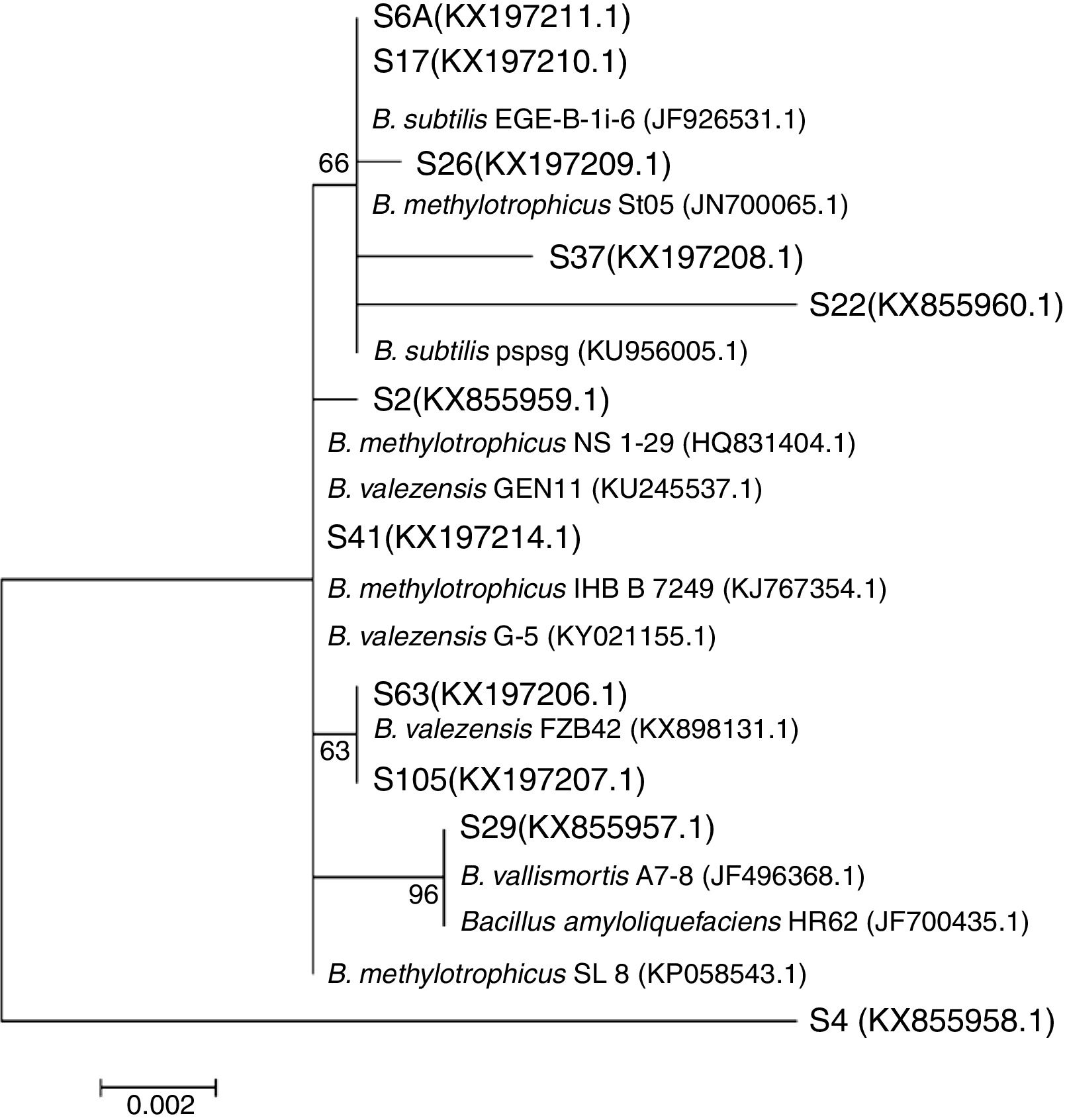
The phylogeny analysis considering only the genus Bacillus showed that the isolates S6A, S17, S26, S37 and S22 are closely related with the strains EGE-B-Li-6 and St05 of B. subtilis and B. methylotrophicus, respectively, while the isolates S2 and S41 are closely related with different strains of B. methylotrophicus and B. valezensis. On the other hand, the isolates S63 and S105 clustered with the strain FBZ42 of B. valezensis. Also, the isolate S29 showed high similarity with B. vallismortis and B. amyloliquefaciens. This analysis also revealed that the isolate S4 is an out group, showing low similarity with the different strains of the genus Bacillus. Even belonging to Bacillus genus (Fig. 2) and showing 98% of identity on the BLAST analysis with the strain SL 8 of B. amyloliquefaciens, the specie of this isolate is still uncertain needing further studies to its determination (Fig. 3).
Based on the results of the phylogenetic analysis, the isolates S4, S22, S29 and S105 were chosen to be evaluated as plant growth promoters under greenhouse conditions. Results showed a clear effect of the isolates on the root development of upland rice plants cv. Aimoré (Fig. 4).
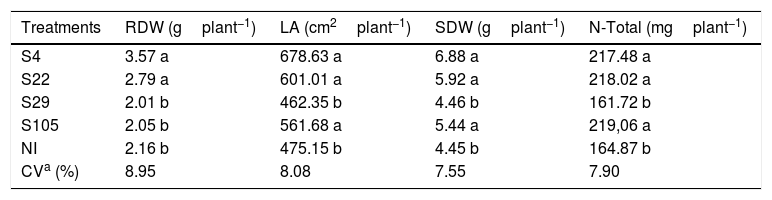
The plant growth promotion ability of the isolates was also observed over root dry weight (RDW), leaf area (LA), shoot dry weight (SDW) and total N (N-Total) accumulated on the shoots of rice plants as observed in Table 4. The isolates S4 and S22 promoted higher values of RDW, LA, SDW and N-Total as compared to NI treatment (Table 4).
Effect of different isolates on the leaf area (LA), root dry weight (RDW), shoot dry weight (SDW) and total N (N-Total) of upland rice cv. Aimoré as compared to a no-inoculated treatment (NI). Mean values of four replicates.
| Treatments | RDW (gplant−1) | LA (cm2plant−1) | SDW (gplant−1) | N-Total (mgplant−1) |
|---|---|---|---|---|
| S4 | 3.57 a | 678.63 a | 6.88 a | 217.48 a |
| S22 | 2.79 a | 601.01 a | 5.92 a | 218.02 a |
| S29 | 2.01 b | 462.35 b | 4.46 b | 161.72 b |
| S105 | 2.05 b | 561.68 a | 5.44 a | 219,06 a |
| NI | 2.16 b | 475.15 b | 4.45 b | 164.87 b |
| CVa (%) | 8.95 | 8.08 | 7.55 | 7.90 |
Values followed by the same letter within each column are not significantly different by the Skott–Knott test (p<0.05).
The morphological analysis revealed four different morphological groups with the prevalence of isolates showing irregular shape, wrinkled border and Gram+ bacteria as well (Table 1). The genus Bacillus sp. comprises Gram+ bacteria and several species of this genus show colony morphology quite similar to those found in this study.29
All isolates were able to use sucrose and malic acid as a carbon source (Table 2). The ability to metabolize sucrose indicates that these isolates have enzyme invertase, which catalyses the hydrolysis of sucrose, allowing those isolates to use this compound as an energy source.30 Malate, the ionized form of malic acid, is a second preferred carbon source for B. subtilis.31
We also found the enzyme amylase in most of the isolates (Table 2). Amylases are widely distributed in bacteria and fungi and are classified by the action of exoenzymes, endoenzymes and enzyme debranching.4
Bacteria utilize citrate through the enzyme citrate lyase. In addition, those bacteria can produce other organic acids derivatives, which can act chelating aluminum and phosphorus molecules.32 In our study, among the positive bacteria for citrate lyase only one did not show the capacity to solubilize inorganic phosphate, indicating that for the other bacteria a synergy between these activities may occur.
Among the isolates, ten of them showed activity for nitrate reductase and nitrogenase, and three for urease activity (Table 2). These enzymes are determining factors related to biological nitrogen fixation and nitrogen metabolism.2 The presence of these enzymes indicates that these isolates show metabolic pathways involved in N assimilation and, thus they can contribute to plant growth.7
On the other hand, only two isolates did not show inorganic phosphate solubilization capacity (Table 3). Besides, among the bacteria studied in this study, the highest inorganic phosphate solubilization index (SI) was about 134%. Several soil bacteria can solubilize inorganic phosphate and provide P for plant growth, increasing P utilization efficiency.33 Bacteria like Pseudomonas and Acetobacter can show SI of 260% and 483%, respectively.34
On studies with bacteria isolated from Brazilian soils, the average SI was 170.5% and the bacteria LMG1222, which belongs to the genus Burkholderia was the most efficient, with a SI of 251%.35 The mean SI observed for the tested isolates in this study was 122.9%, with the highest values around 133% for isolates S2 and S5 (Table 3).
Another important tool for plant growth-promoting bacteria is their ability to produce proteases. The production of protease by bacteria is often studied and has great importance for industrial use and on the understanding of the activities occurring in the soil environment.3 According to Oliveira et al.,13 bacteria showing protease enzymatic index (PEI) ≥2 are considered good protease producers.
Our results revealed five good protease producers bacteria (Table 3). Isolate S6 showed PEI about 3, which is very close to the PEI values showed by Bacillus megaterium and Corynebacterium renale (3.1 and 4.3, respectively) isolated from Jacaranda decurrens plants, isolated from soils of the state of Goiás, Brazil.36
The enzymatic diversity displayed by endophytic bacteria isolated from upland rice grown in Cerrado indicates that it is possible to develop a strategy to benefit the selection of microorganisms as biofertilizers, improving their potential as plant growth promoters for rice crop. Thus, the greater the extent of enzymatic pathways presented by the microorganisms, the greater will be the generated benefit. Also, the success of the plant–bacterium interaction and competition with other microorganisms favors the establishment of those microorganisms in such environments.3
The ability of endophytic bacteria strains to grow using carbon source present in soil and plants as substrate indicates their ability to survive and compete in the environment. Organic acids such as maleic and nicotinic acid can also be exuded by the roots of plants, playing an important role in the selection of the associated microbiota. Nicotic and maleic acids showed suppressive effect over most of the studied isolates (Table 2), which corroborates the findings of Fernandes Junior et al.4
Among the evaluated bacteria, 55% and 70% showed antibiosis against S. sclerotiorum and R. solani, respectively (Table 3). Antibiosis may be triggered by many processes, including competition and production of metabolites.37 Isolates S4, S6, S22, S25, S29 and S105 showed antibiosis, solubilization of inorganic phosphate and protease activities (Table 3). According to Park et al.,38 the solubilization of inorganic phosphate and protease activity may be involved in the antibiosis activity of bacteria against fungi. These bacteria showed a very close similarity based on their morphological and enzymatic activity, carbon source use and antibiosis data. Isolates S4, S6, S25 and S29 shared about 62% of similarity on group G2; isolates S22 and S105 about 65% of similarity on group G4. The similarity between groups G2 and G4 was about 52% (Fig. 1).
Most of the isolates evaluated in this study, especially S4 and S22, show different mechanisms involved in the plant growth promotion, besides the presence of enzymes activity related to adaptation to different environmental conditions, as those of Cerrado. In this way, the results of this study represent a great scientific contribution since many metabolic pathway and action mechanisms can be studied from a few number of isolates. Besides, the contribution from the agricultural point of view is highly promising for the Brazilian Cerrado because there are none biological products registered for the upland rice crop.
The phylogenetic analysis of the 16S rRNA gene sequencing, performed for 11 isolates revealed that all of them belong to the genus Bacillus (Fig. 2). The isolate S4 showed low similarity with the different Bacillus spp., remaining as an undefined specie (Fig. 3). The presence of the genus Bacillus associated to rice plants in Brazil was also reported,39 but never with the presence of the B. amyloliquefaciens species, as observed in this study. In a study that investigated the associated microbiota with tomato, the presence of B. amyloliquefaciens was related with antibiosis against pathogens.40 However, many other factors, such as soil type and plant genotype may influence this plant–microorganism association.39,41
In our study, the best results for plant growth promotion under greenhouse conditions were observed for the isolates S4 and S22 (Table 4). As reported before, these isolates show different traits related to plant growth promotion, such as antibiosis, solubilization of inorganic phosphate and protease activity. The isolate S22 increased root dry weight, leaf area, shoot dry weight and N-Total accumulated on rice shoots in about 23%, 21%, 25% and 24%, respectively, as compared to noninoculated treatment. Considering these same parameters, the isolate S4, an unknown specie, promoted an increase of about 40%, 30%, 35% and 24%, respectively (Table 4).
The Brazilian Cerrado is a very challenging environment for cultivated plants, like rice, due to its soil and climate characteristics. Cerrado soils are generally acid and poor in nutrients,42 while the rainfall regime is well defined, with a rainy season from October to April and a dry season from May to September with an average annual rainfall of 1460mm.43 Then, the use of microorganisms as inoculant showing a great variability of traits related with the growth promotion of plants is a very important approach to improve the establishment of rice crops in the Brazilian Cerrado, since plants under influence of PGPR can be greater, stronger, more productive and healthier.3,5,6
These results point out for further studies on the specie identification of the isolate S4. Also, the isolates S4 and S22 must be tested for their agronomic efficiency under field conditions, aiming to state their potential of use as inoculant for rice.
ConclusionsThis study reveals great morphological, metabolic and genetic variability among bacterial isolates obtained from upland rice cultivated in the Cerrado region of the state of Goiás. Those bacteria show many enzymes activity related to adaptation to different environmental conditions, as those of Cerrado. All the isolates belong to the genus Bacillus and exhibit at least one mechanism associated to plant growth promotion. Under greenhouse conditions the isolates S4 and S22 showed high plant growth promotion, indicating that they should be tested for this ability under field conditions.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank CAPES, Embrapa Rice and Beans and INCT on Biological Nitrogen Fixation for the financial support. The Department of Biochemistry and Molecular Biology at the Federal University of Paraná and Goiás State University for researcher exchange (BIP).
Part of the Master's Dissertation of the first author approved by the Molecular Science Post-Graduation Program of the Goiás State University.