Polychlorinated biphenyls (PCBs), the chlorinated derivatives of biphenyl, are one of the most prevalent, highly toxic and persistent groups of contaminants in the environment. The objective of this study was to investigate the biodegradation of PCBs in northeastern (Heilongjiang Province), northern (Shanxi Province) and eastern China (Shanghai municipality). From these areas, nine soil samples were screened for PCB-degrading bacteria using a functional complementarity method. The genomic 16S rDNA locus was amplified and the products were sequenced to identify the bacterial genera. Seven Pseudomonas strains were selected to compare the capacity of bacteria from different regions to degrade biphenyl by HPLC. Compared to the biphenyl content in controls of 100%, the biphenyl content went down to 3.7% for strain P9-324, 36.3% for P2-11, and 20.0% for the other five strains. These results indicate that a longer processing time led to more degradation of biphenyl. PCB-degrading bacterial strains are distributed differently in different regions of China.
Polychlorinated biphenyls (PCBs), the chlorinated derivatives of biphenyl, are highly toxic environmental pollutants that have become ubiquitous throughout the environment and the food web because of their low degradability. Therefore, the removal of PCBs from soil and sediment is a high priority in several industrial countries.1 PCB molecules consist of a biphenyl nucleus carrying 1–10 chlorines, which can create >200 possible congeners that differ in chlorine number and position. The less-chlorinated congeners are usually less toxic than the more-chlorinated congeners.2 The remediation of PCB contamination in the environment has become extremely important.
The microbial degradation of PCBs is one of the most cost-effective and energy-efficient methods for removing them from the environment. Many PCB-degrading bacteria have been isolated, such as Gram-negative strains belonging to the genera Pseudomonas, Alcaligenes, Achromobacter, Burkholderia, Acinetobacter, Comamonas, Sphingomonas and Ralstonia, and Gram-positive strains of the genera Arthrobacter, Corynebacterium, Rhodococcus and Bacillus.3 The genes encoding the enzymes for biphenyl degradation are clustered and termed bph gene clusters.4 The enzymes can be divided into four types, including biphenyl dioxygenase (BphA), dihydrodiol dehydrogenase (BphB), 2,3-dihydroxybiphenyl dioxygenase (BphC) and hydrolase (BphD).5–11
We previously investigated the distribution of PCB-utilizing bacteria in Shanghai.12 In this study, we investigated the distribution of PCB-utilizing bacteria from other different areas of China. Moreover, we chose several Pseudomonas strains from different soil samples and measured their degradation of biphenyl using HPLC.
Materials and methodsChemicals and mediaPCB (2,2′,3,3′-tetrachlorobiphenyl, CAS No. 38444-93-8) (99.5% purity) and biphenyl (CAS No. 92-52-4) (99.5% purity) were purchased from the J&K Chemical Co Ltd (Shanghai, China) and Shanghai Chemical Agent Co Ltd (Shanghai, China), respectively. The other chemicals used in this study were of the highest purity available. Mineral salt medium (MM)13 contained (g): NH4NO3 1.0, KCl 0.7, KH2PO4 2.0, NaHPO4 3.0, MgSO4·7H2O 0.7, CaCl2 0.02 and NaCl 1.0 in 1L of double-distilled water at pH 7.0. Solid MM was MM containing agar at 16g/L. After autoclaving, tetrachlorobiphenyl was added as the sole carbon source.
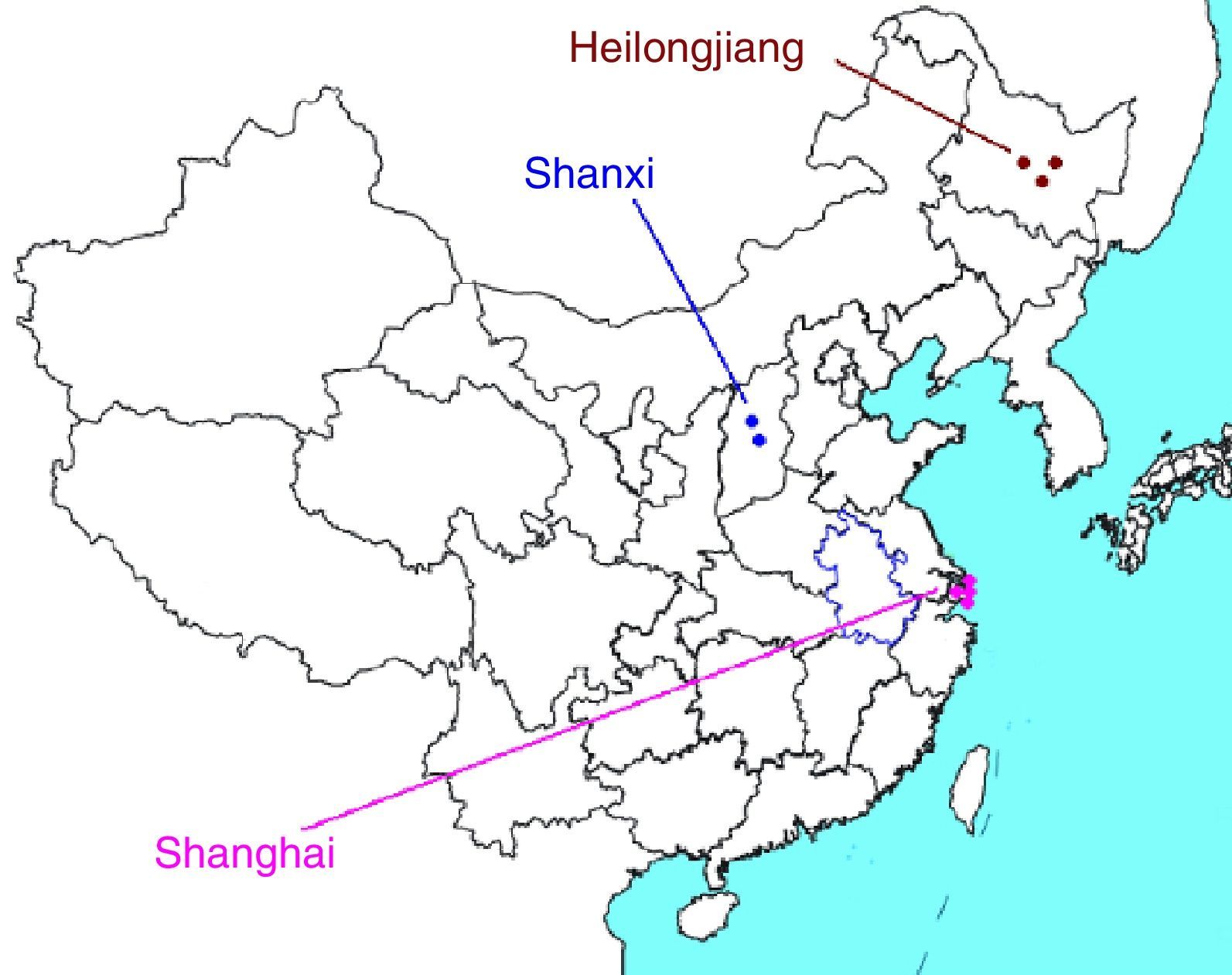
Isolation of PCB-degrading bacterial strainsSamples 1–3 were collected from Heilongjiang Province in northeast China: Sample 1 from a chemical factory; Sample 2 from a botanical garden; and Sample 3 from a pharmaceutical factory. Samples 4 and 5 were collected from Shanxi Province in northern China: Sample 4 from the Fen River and Sample 5 from a paper mill. Samples 6–9 were collected from Shanghai Municipality in eastern China: Sample 6 from an experimental field; Sample 7 from a riverbed not far from an old chemical factory; Sample 8 from an old chemical factory; and Sample 9 from a riverbed near a residential area (Fig. 1).
For each sample, 10.0g of soil was put into a sterilized mortar with 50mL of sterile water, crushed with a sterilized pestle, and then left for 5min. A sample (1–5mL) of the clear supernatant liquid was added to an Erlenmeyer flask that contained 100mL of MM with 0.01% tetrachlorobiphenyl as the sole carbon source. Then, the Erlenmeyer flask was incubated for 3d at 28°C in darkness with shaking at 150rpm. A 1mL aliquot was transferred to another 100mL of fresh MM containing 0.01% tetrachlorobiphenyl and incubated under the same conditions. This process was repeated three times. Of the suspension, 20μL was plated onto solid MM containing 0.01% tetrachlorobiphenyl as the sole carbon source and incubated for 3d at 28°C.
Identification and analysis of 16S rDNA sequencesGenomic DNA was extracted from the PCB-utilizing bacterial strains.14 The 16S rDNA locus was amplified using two primers (5′-ACG GCTACCTTGTTACGACTTC-3′ and 5′-AGAGTTTGATCCTGGCTCAG-3′) and Ex-taq polymerase with genomic DNA as the template, separated by 1.5% agarose gel electrophoresis, and cloned into the pMD-18 vector (TaKaRa, Dalian, China) for enzymatic and sequencing identification. The PCR protocol was 94°C for 10min, then 35 cycles of 94°C for 30s, 50°C for 30s, 72°C for 2min, and a final extension at 72°C for 10min in a 50μL reaction volume. The PCR products were separated by 1.5% agarose gel electrophoresis and quantified using a Model Gel Doc 1000 analyzer (Bio-Rad, Hercules, CA). Phylogenetic analysis was based on the 16S rDNA by MEGA5 software and the neighbor-joining (NJ) method with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates).15
HPLC analytical assay of biphenyl degradationTo verify and compare the extent of degradation, we chose seven Pseudomonas strains: P1-5, P2-11, P3-24, P4-38, P5-31, P7-245 and P9-324 from seven soil samples. The details of detecting biphenyl degradation by HPLC were as follows: each of the seven strains (100μL) and a control (no strain) were cultured in an Erlenmeyer flask (500mL) containing 100mL of LB medium at 28°C in a rotary shaker at 120rpm overnight. The next day, 1mL of biphenyl solution (50mg/mL, dissolved in methyl alcohol) was added into each Erlenmeyer flask and the culture was continued. After 2 and 4d, 500μL of each solution was placed into an Eppendorf tube and then 500μL of ether was added. The tubes were deposited on an oscillator for 5–10min so that the biphenyl was fully extracted by the ether. The supernatant of the two tubes was withdrawn and mixed together in a new tube, 500μL of methyl alcohol was added and the mixture was ready for rotary evaporation. The ether was fully evaporated and the biphenyl was kept in the methyl alcohol to keep it from adhering to the wall of the round-bottomed bottle.12
Before HPLC analysis, each sample was membrane-filtered. A 10-μL sample of the resulting solution was subjected to reverse-phase HPLC (1100 series, Agilent Co, USA). The separation column for the HPLC (4.6mm×150mm×5μm) was filled with Kromail 100-5C18. The mobile phase contained acetonitrile/water (70:30, v/v) and the flow rate was 1.0mL/min. Biphenyl was detected at a wavelength of 254nm.4 A solution of pure biphenyl (100μL/mL) was used to determine the peak retention time.
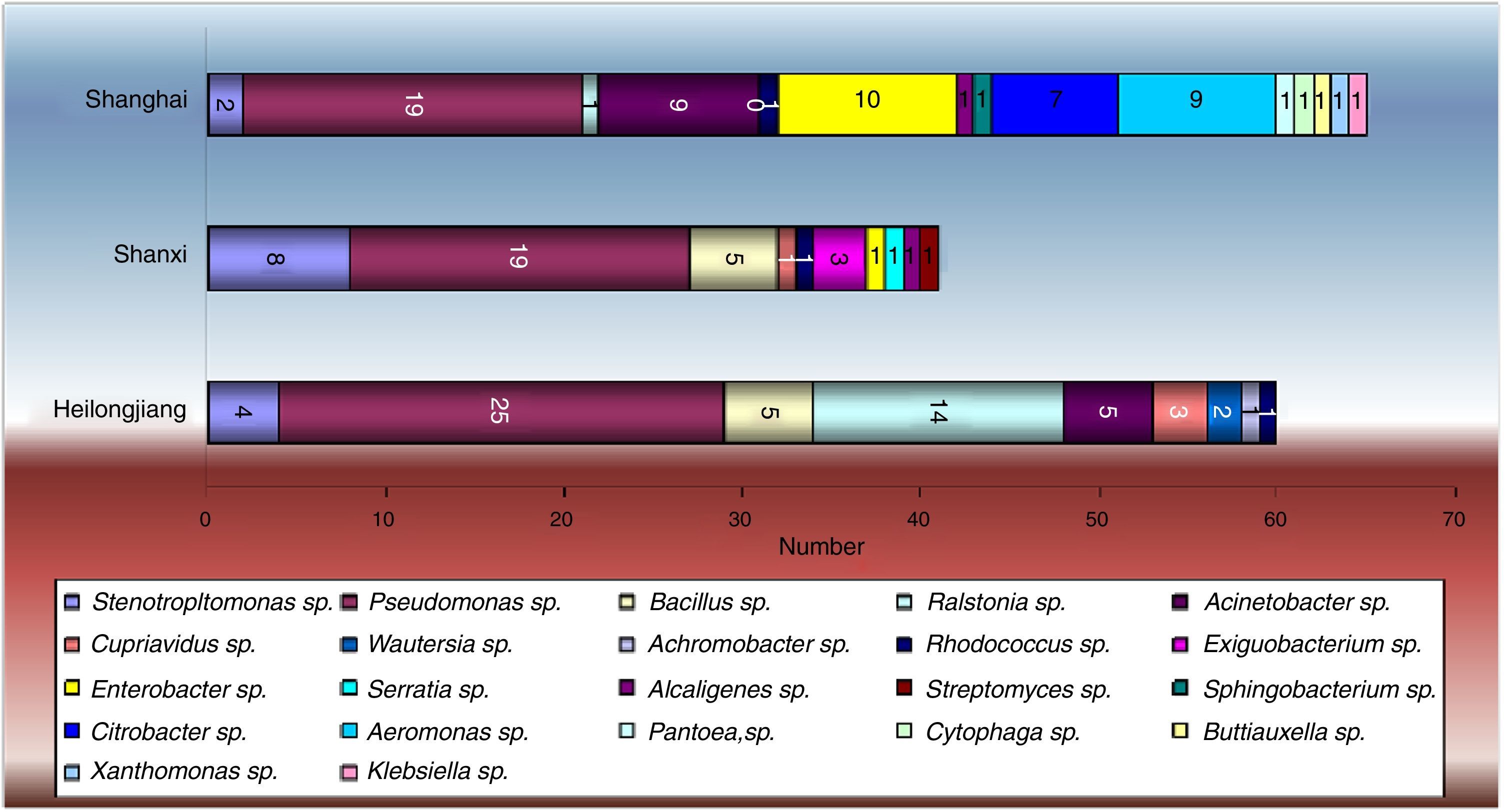
ResultsIsolation and classification of PCB-degrading bacteriaFrom all the soil samples, a total of 22 PCB-utilizing bacterial strains were detected, including the following genera: Stenotrophomonas, Pseudomonas, Bacillus, Ralstonia, Acinetobacter, Cupriavidus, Wautersia, Achromobacter, Rhodococcus, Exiguobacterium, Enterobacter, Serratia, Alcaligenes, Streptomyces, Sphingobacterium, Citrobacter, Aeromonas, Pantoea, Cytophaga, Buttiauxella, Xanthomonas and Klebsiella (Fig. 2). There were 66 strains identified from Shanghai, more than 60 from Heilongjiang, and 45 from Shanxi. Furthermore, the strains from Shanghai encompassed 15 categories, more than the other two regions. Some bacterial strains were found in all three regions, e.g., Stenotrophomonas, Pseudomonas and Rhodococcus. However, some bacteria only existed in a particular soil, e.g., Achromobacter was only in soil samples from Heilongjiang; Exiguobacterium, Serratia and Streptomyces were only in soil samples from Shanxi; Sphingobacterium, Citrobacter, Aeromonas, Pantoea, Cytophaga, Buttiauxella, Xanthomonas and Klebsiella were only in soil samples from Shanghai.
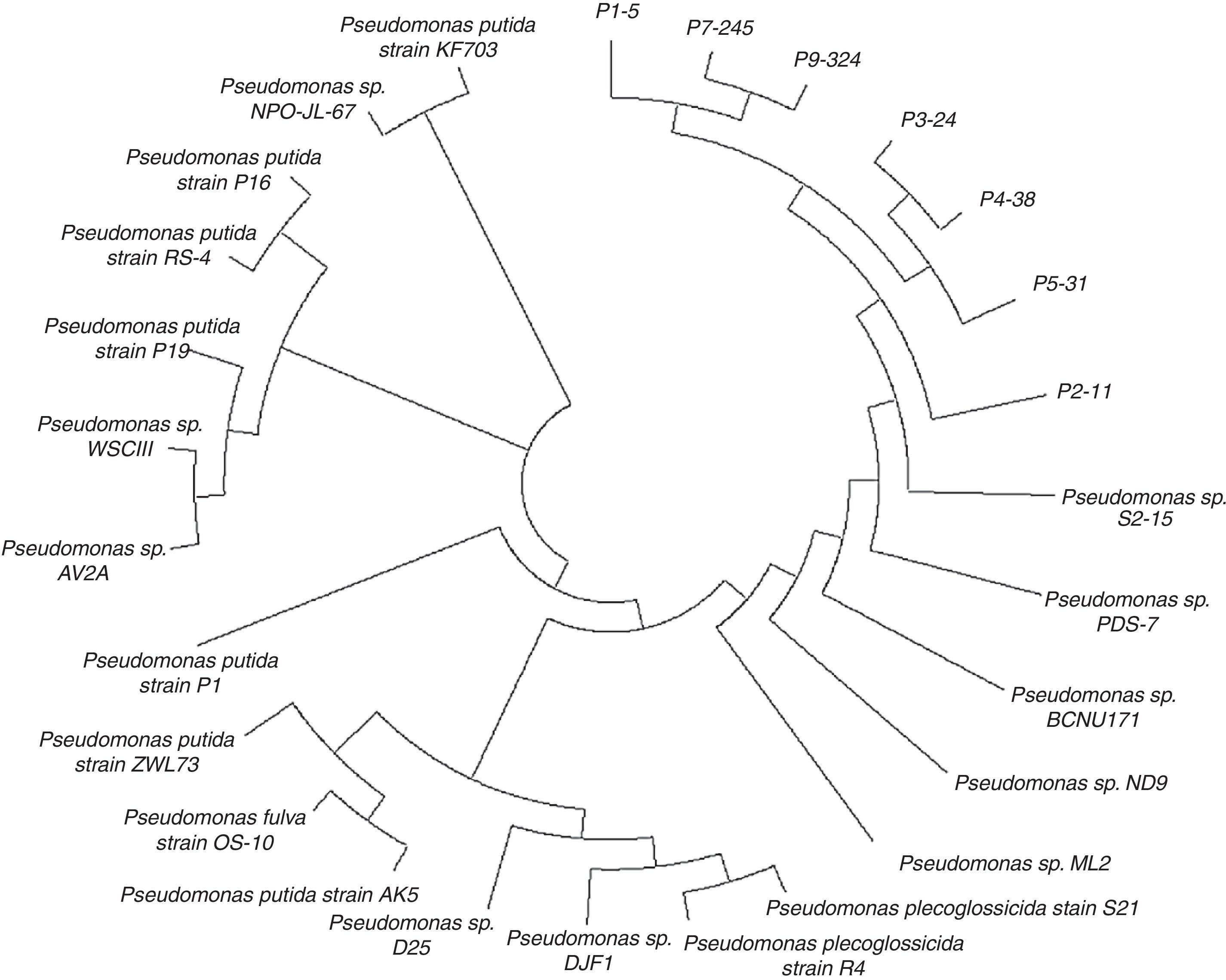
Phylogenetic reconstruction of the selected strainsA multiple alignment analysis using full 16S rDNA sequences was used to study the phylogenetic relationships among the seven Pseudomonas strains selected from different soil samples and other Pseudomonas strains. Strains P7-245 and P9-324 were found to belong to one branch, and P3-24 and P4-38 belong to a separate branch, with each of these showing the closest relationship (Fig. 3). Moreover, strains P1-5 and P5-31 occupy, large sub-branches of the two above-mentioned branches. Strain P2-11 is in a different branch from the other six Pseudomonas strains.
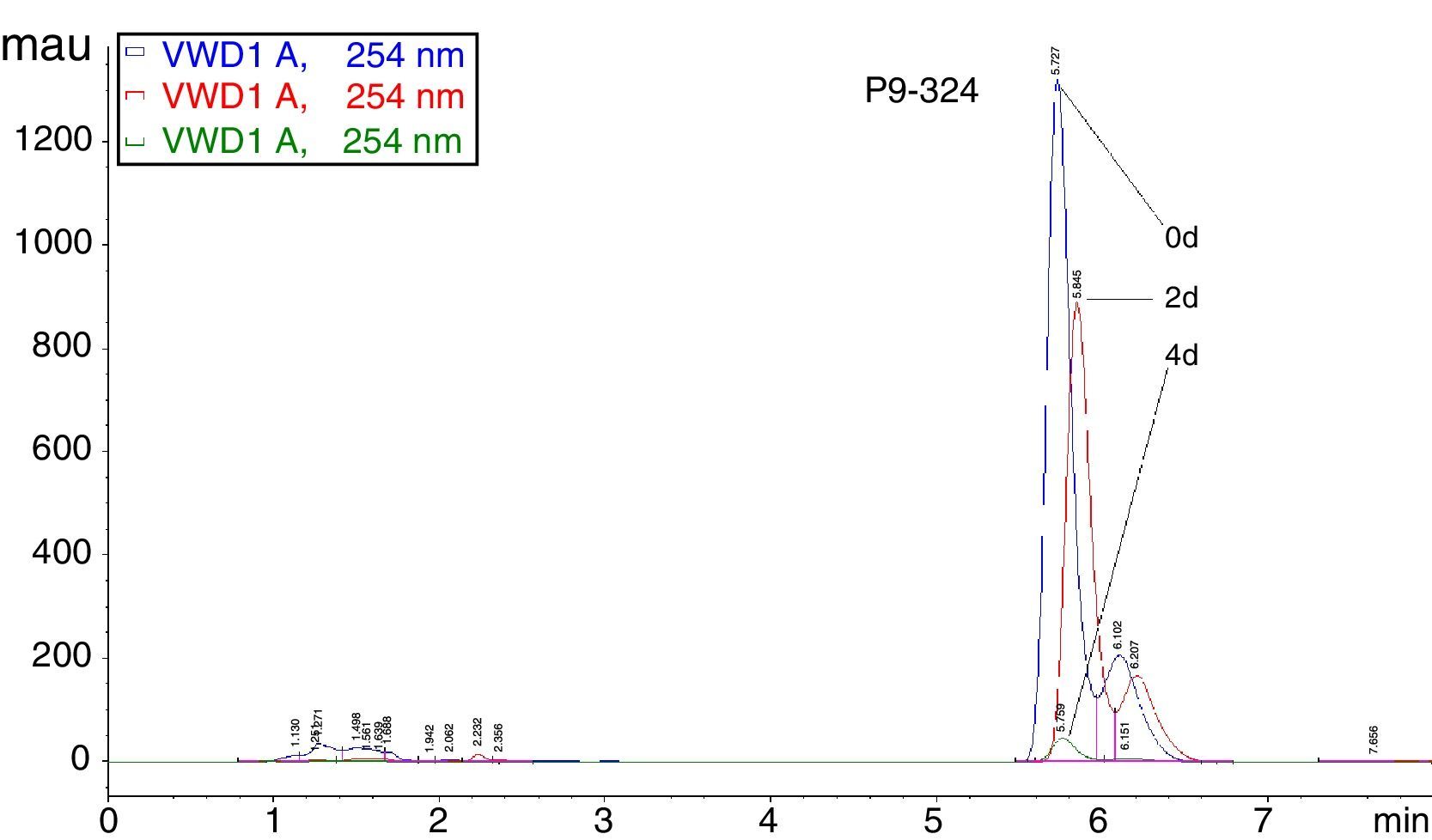
Degradation of biphenylHPLC was used to detect the efficiency of the seven Pseudomonas strains at biphenyl degradation. When pure biphenyl was subjected to HPLC, the highest peak appeared at a retention time of ≈5.8min, indicating the point of biphenyl detection.12 The peak's area is directly proportional to the sample concentration. In our assay, there was only one large peak.
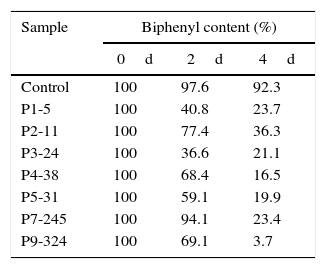
After 2d, assay results showed that each strain could degrade biphenyl with the degradation rate ranging from 5.9% (strain P6-245) to 65.3% (strain P4-38) (Table 1). After 4d, strain P9-324 had degraded the maximum amount of biphenyl (96.3%), whereas the other strains all degraded amounts varying from 63.7% (strain P2-11) to 83.6% (strain P4-38) (Fig. 4). However, the amount of biphenyl in the controls had changed little by either 2 or 4d.
PCBs refer to a large group of chlorinated biphenyls with 209 possible congeners.16 Due to their hydrophobic properties, PCBs tend to become absorbed by natural organic matter in soil, sediments and sludges. PCBs are a serious environmental problem due to their toxicity and persistence in the environment. Natural biodegradation of PCBs in contaminated soil and sediment occurs at low rates,17 and various attempts to accelerate biodegradation have been made.18–21
Despite active research in developed nations spanning three decades, extensive regulatory action, and an effective ban on their production since the late 1970s, PCBs remain a focus of research and environmental attention.22 China is a developing country in which pollution prevention and control is most likely less effective compared to developed countries. Moreover, PCBs are widespread in the environment and are found globally as a result of ocean currents and atmospheric deposition.23,24 Thus, it is urgent to study PCB contamination and biodegradation in China and across the globe.
The PCB-biodegrading bacterial strains in Shanghai, the biggest industrial metropolis in China, have been characterized in detail.12 The objective of this study was to investigate and compare the PCB-degrading bacteria in northeastern (Heilongjiang Province), northern (Shanxi Province) and eastern China (Shanghai Municipality). Thus, research on PCB contamination and biodegradation in large-scale regions is urgently needed. Three regions were selected to investigate PCB-degrading bacteria. Heilongjiang Province, located in northeast China, is a main agricultural (rice and corn production) region. Shanxi Province, located in northern China, is a main energy (coal production) region. Shanghai has developed rapidly in recent decades and along with growth of the economy, environmental pollution is a subject worthy of concern. Soil samples were collected from northeastern, northern and eastern China to investigate the biodegradation of PCBs. Seven categories of strains were identified in the soil samples from Heilongjiang and Shanxi Provinces but not in the soil samples of Shanghai.
Here, we chose nine soil samples from northeastern, northern and eastern China, based on areas where industry has quickly developed and were thus more likely to have polluted the environment. There were >2 sampling locations in each region to make the results more representative. The PCB-degrading bacterial strains were varied, with strains of genera Stenotrophomonas, Pseudomonas and Rhodococcus present in each region; these three PCB-utilizing genera have been reported in previous studies.25 In contrast, some bacteria were found to exist only in a single region, possibly due to the different geographical locations. Thus, the distribution of PCB-degrading bacterial strains may be closely linked to geographic location, and the state of development in the region may also play a role in the evolutionary selection.
According to the phylogenetic tree (Fig. 3), the relationships among the seven Pseudomonas strains were closer than with the others. HPLC confirmed that the seven Pseudomonas strains could degrade biphenyl. The sampling results at 2d showed that all bacteria had started to degrade biphenyl. By 4d, the degradation of biphenyl by all strains was not the same. Strain P9-324 had degraded nearly 96.0% of the biphenyl compared with the control, and strain P2-11 had degraded 63.7%, which was less than the other six Pseudomonas strains. In addition to strains P9-324 and P2-11, the other five Pseudomonas strains degraded approximately 80.0% of the biphenyl. These results are relatively consistent with the phylogenetic tree. Strain P2-11 came from Sample 2, which was from a botanical garden in Heilongjiang. Botanical gardens are well protected and less susceptible to outside contamination; this balanced ecosystem may cause the resistance of bacteria there to be relatively weaker than in other places, e.g., strain P2-11 2 degraded only 63.7% of the biphenyl. Strain P9-324 isolated from Sample 9 was from a riverbed near residential quarters and had the strongest ability to degrade biphenyl of approximately 96.3%. It is most likely that the river was seriously polluted many years ago, and these strains remained in the mud and retained their strong ability to degrade biphenyl.
In the present study, different areas in China were screened for the presence of PCB-degrading bacteria due to interest in geographical influences. We combined molecular biology and chemistry together in the experiments. Our future work will include searching for the sources of environmental pollution, solving the problems of pollution by biodegradation and combining multi-disciplinary methods to prevent pollution.
Conflict of interestThe authors declare no conflicts of interest
The research was supported by A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).