Chicha, a type of beer made mainly with maize or cassava, is a traditional fermented beverage of the Andean region. There have only been a few studies on yeasts associated with chicha fermentation, and the species diversity occurring during the production of this beverage is not known. The objective of this study was to determine the biodiversity of yeasts in chicha, and to characterize the Saccharomyces cerevisiae populations associated with the production of chicha de jora, seven-grain chicha, chicha de yuca, and chicha de morocho in Ecuador. The molecular diversity of S. cerevisiae populations was determined by restriction polymorphism mitochondrial profiles. The beverages were characterized based on their physicochemical parameters. Twenty-six species were identified, and the most prevalent species were S. cerevisiae and Torulaspora delbrueckii. Other yeast species were isolated at low frequencies. Among 121 isolates of S. cerevisiae, 68 different mtDNA molecular profiles were identified. These results showed that chichas are fermented by a high number of different strains of S. cerevisiae. Some other species provided a minor contribution to the fermentation process. The chicha presented generally similar physicochemical parameters to those observed for other traditional fermented beverages, and can be considered as an acid fermented beverage.
Chicha or maize beer could be regarded as the oldest beverage in Latin America. The name chicha possibly originates from the word chichab, from the original language spoken in the current territory of Panama, which means maize. Other theories suggest that the name is derived from the word Chibcha, a civilization that populated Colombia and Panama, or relate the word chicha to Chichas, an ethnicity present in southern Bolivia before the establishment of the Incas.1
Chicha is a clear, yellow, and frothy beverage present in the Andean region and in low-lying regions of Ecuador, Peru, Bolivia, Colombia, Brazil, and Argentina.2 This traditional beverage is prepared mainly from maize, but currently, the name is considered generic and refers to a variety of beverages, fermented or not, prepared from various other materials, such as cassava, beans (such as rice, oats, and quinoa) and fruits (such as bananas). In Ecuador, the first reports of chicha production date back to 200 B.C., before the establishment of the Incas in the region.1 This beverage was of great importance in traditional indigenous cultures, especially in the Incan culture, wherein it was also linked to festive ceremonies.3
In Ecuador, as in the rest of the Andean region, the most common maize chicha is chicha de jora (Fig. 1). This chicha is prepared from the yellow maize grain (maíz amarillo), which is malted (germinated and dried). For the preparation of malt, the grains are left in water for a day. This step is necessary to achieve the optimum grain moisture for germination. Subsequently, the water is drained and the maize is placed in baskets of stray to germinate over a period of 13 days. Once germinated, the maize is put into straw mats or plastic tarps under the sun for 2 days to dry completely, which stops the enzymatic activity within the grain. After drying, the beans are ground and the flour obtained is used for the preparation of chicha. For this, the jora flour is added to cold water and then this mixture is transferred to vessels with hot water, and boiled for approximately 20min. After boiling, the mixture is strained and then placed in a container to ferment. The clay vessels, formerly used for boiling and fermentation, have been replaced by aluminium pots and plastic containers, respectively. The spent grain obtained after filtration is termed afrecho and serves as food for animals. The fermentation vessels are usually open. Usually after two days of spontaneous fermentation by indigenous microorganisms, the beverage is ready for consumption. Some producers typically boil jora flour with other ingredients, including panela (brown sugar in solid pieces). Others make a mixture of panela and herbs and then add this mixture to the jora flour and water. There are still those that add pieces of fruit and panela to the beverage, after filtering.
Other chicha beverages produced in Ecuador include chicha de morocho, made with white maize, and chicha produced with seven varieties of maize including jora, maíz amarillo (yellow maize), maíz blanco (white maize), maíz negro (black maize), chulpi (chulpi maize), morocho (morocho maize) and cangil (popcorn maize). Seven-grain chicha is produced in the town of Otavalo, in northern Ecuador, and is a very famous drink and appreciated throughout the country. The yuca (cassava, Manihot esculenta) is also an important raw material for the production of chicha.4 This chicha is produced by the indigenous and mestizo population in the Amazon region of Ecuador.
Few studies have been performed to identify the yeast species in chichas. Vallejo et al.5 isolated Saccharomyces cerevisiae as the single yeast species at the end of fermentation from 10 samples of chicha de jora collected from 10 different familiar traditional “chicherías” in the Cusco region of Peru. These authors suggested that this species was mainly responsible for alcoholic fermentation in these chicha samples. Rodríguez et al.6 suggest that Saccharomyces uvarum is responsible for the traditional fermentation of apple chicha elaborated by aboriginal communities of Andean Patagonia (Argentina and Chile). Mendonza et al.7 showed by high-throughput sequencing and culture-dependent approaches that S. cerevisiae was the dominant species in an Argentinian maize-based chicha. Other works on chicha fermentation linked bacterial populations to this process.4,8,9 Despite the work of Vallejo et al.5 and Mendonza et al.,7 the yeast biodiversity associated with maize and cassava chicha production is almost unknown. In this work, chichas sold in bulk (Fig. 1), produced with different substrates and different fermentation times, were collected from markets, bars, restaurants, and in villages of Ecuador. The objective was to determine yeast species richness and to characterize the S. cerevisiae populations associated with the production of this beverage by restriction polymorphism mitochondrial DNA (mtDNA) analyses. In addition, the physicochemical parameters of the beverages were determined.
Materials and methodsSamplingForty-two chicha samples were collected from August to October of 2010 and April to September of 2012 in two regions of Ecuador: the Amazon region, within the Yasuní National Park (Orellana Province) and the Andean region, in the provinces of Pichincha, Imbabura and Chimborazo. The samples included two chichas de yuca, 34 chichas de jora, three seven-grain chichas, and two chichas de morocho. In these samples, the fermentation was considered finished by the producers, and the beverage was ready to drink. One sample of chicha de jora was sampled during successive fermentation times (0–5 days). The chichas were collected in sterile bottles of 100mL, transported to the laboratory on ice, and processed the same day.
Yeast isolation and identificationAliquots of 25mL of each chicha were added to 225mL of sterile 0.1% peptone water. For yeast isolation, 0.1mL of appropriate decimal dilutions, in triplicate, was spread on yeast extract-malt extract (YMA: 1% glucose, 0.5% peptone, 0.3% malt extract, 0.3% yeast extract, 2% agar, and 0.02% chloramphenicol) and lysine (1.17% YCB, 0.056% lysine, 2% agar, and 0.02% chloramphenicol) agars. The YMA was utilized for the isolation of Saccharomyces and non-Saccharomyces yeasts while the lysine agar was utilized for the isolation of non-Saccharomyces yeasts. The plates were incubated at 25°C for 5 days and the density of each different yeast morphotype was expressed as the means of each morphotype in each sample in colony-forming units (cfu/mL). Representative colonies of each different yeast morphotype from both culture media were purified on YMA plates and preserved at −80°C or in liquid nitrogen for subsequent identification.
The yeast isolates were morphologically and physiologically characterized according to Kurtzman et al.10 Isolates with identical morphological and physiological characteristics were grouped together and subjected to PCR using the core sequences of the primer (GTG)5 as described by Gomes et al.11 Isolates with identical DNA banding patterns were grouped and tentatively considered the same species. At least 50% of the yeast isolates of each molecular group were identified by sequencing. Species identification was performed by sequence analysis of the ITS-5.8S region and the D1/D2 variable domains of the large subunit of rRNA gene as described previously.12–16 Sequencing was performed directly from purified PCR products using an ABI3130 (Life Technologies, USA) automated sequencing system. For yeast species identification, the sequences obtained were compared to those included in the GenBank database using the Basic Local Alignment Search Tool (BLAST at http://www.ncbi.nlm.nih.gov).
Restriction polymorphism of mitochondrial DNAExtraction of mtDNA from 121 isolates of S. cerevisiae was performed according to the methodology described by Querol and Barrio17 and modified by Foschino et al.18 The mtDNA was resuspended in 25μL of TE and stored at −20°C. For digestion, a mixture containing 10× buffer (Invitrogen, USA), 1μL of RNAse A (Invitrogen, USA), 1μL of Hinf I (Invitrogen, USA), and 10μL of DNA (approximately 1500ng) was used. The volume was completed with deionized water to 20μL. The tubes were then incubated at 37°C for 6h. The mtDNA restriction fragments were separated by electrophoresis on a 1.5% agarose gel (Pronadisa, Spain) in TBE buffer at 80V for 2h. The gels were stained with a solution of GelRed (Biotium, USA) and visualized and photographed under ultraviolet (UV) light.
Physicochemical analysesThe parameters of pH, total reducing sugars, and ethanol content were determined. The determination of total reducing sugars was performed by the 3,5-dinitrosalicylic acid methodology.19 The ethanol, glycerol, and lactic and acetic acid contents were determined by high performance liquid chromatography (HPLC) in an Agilent chromatographer model equipped with a column Rezex ROA (300×7.8mm).
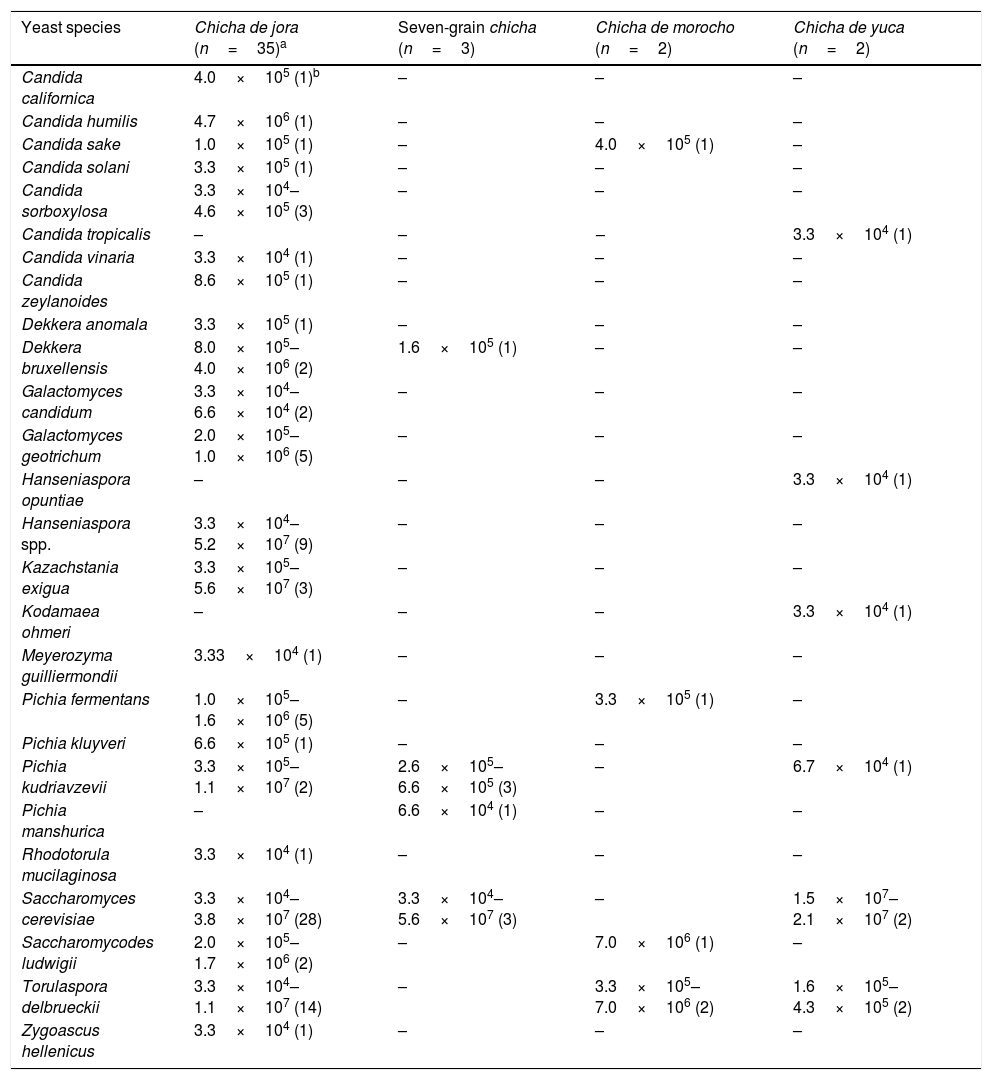
ResultsYeast identificationTwo hundred and fifty-four yeast isolates obtained from chicha samples were identified as belonging to 26 species. S. cerevisiae was the most prevalent species occurring in 33 of 42 chicha samples (41 samples plus one sample collected in different time), followed by Torulaspora delbrueckii that was identified in 18 samples. These two species as well as Pichia kudriavzevii, Candida sake, Dekkera bruxellensis, Pichia fermentans, and Saccharomycodes ludwigii were isolated from at least two types of chicha (Table 1). The other yeast species were present in only one type of chicha. The highest yeast counts were obtained for chicha de jora and seven-grain chicha. S. cerevisiae populations contributed to the highest yeast counts in most samples of chichas de jora, seven-grain chichas, and chichas de yuca. In most samples that this species was detected, the population counts were above 1.0×107cfu/mL. However, this species was not isolated from two samples of chicha de morocho, and T. delbrueckii and S. ludwigii presented the highest counts (7.0×106cfu/mL) in this beverage.
Yeast species (cfu/mL and number of positive samples for each species) isolated from chicha samples collected in Ecuador.
| Yeast species | Chicha de jora (n=35)a | Seven-grain chicha (n=3) | Chicha de morocho (n=2) | Chicha de yuca (n=2) |
|---|---|---|---|---|
| Candida californica | 4.0×105 (1)b | – | – | – |
| Candida humilis | 4.7×106 (1) | – | – | – |
| Candida sake | 1.0×105 (1) | – | 4.0×105 (1) | – |
| Candida solani | 3.3×105 (1) | – | – | – |
| Candida sorboxylosa | 3.3×104–4.6×105 (3) | – | – | – |
| Candida tropicalis | – | – | – | 3.3×104 (1) |
| Candida vinaria | 3.3×104 (1) | – | – | – |
| Candida zeylanoides | 8.6×105 (1) | – | – | – |
| Dekkera anomala | 3.3×105 (1) | – | – | – |
| Dekkera bruxellensis | 8.0×105–4.0×106 (2) | 1.6×105 (1) | – | – |
| Galactomyces candidum | 3.3×104–6.6×104 (2) | – | – | – |
| Galactomyces geotrichum | 2.0×105–1.0×106 (5) | – | – | – |
| Hanseniaspora opuntiae | – | – | – | 3.3×104 (1) |
| Hanseniaspora spp. | 3.3×104–5.2×107 (9) | – | – | – |
| Kazachstania exigua | 3.3×105–5.6×107 (3) | – | – | – |
| Kodamaea ohmeri | – | – | – | 3.3×104 (1) |
| Meyerozyma guilliermondii | 3.33×104 (1) | – | – | – |
| Pichia fermentans | 1.0×105–1.6×106 (5) | – | 3.3×105 (1) | – |
| Pichia kluyveri | 6.6×105 (1) | – | – | – |
| Pichia kudriavzevii | 3.3×105–1.1×107 (2) | 2.6×105–6.6×105 (3) | – | 6.7×104 (1) |
| Pichia manshurica | – | 6.6×104 (1) | – | – |
| Rhodotorula mucilaginosa | 3.3×104 (1) | – | – | – |
| Saccharomyces cerevisiae | 3.3×104–3.8×107 (28) | 3.3×104–5.6×107 (3) | – | 1.5×107–2.1×107 (2) |
| Saccharomycodes ludwigii | 2.0×105–1.7×106 (2) | – | 7.0×106 (1) | – |
| Torulaspora delbrueckii | 3.3×104–1.1×107 (14) | – | 3.3×105–7.0×106 (2) | 1.6×105–4.3×105 (2) |
| Zygoascus hellenicus | 3.3×104 (1) | – | – | – |
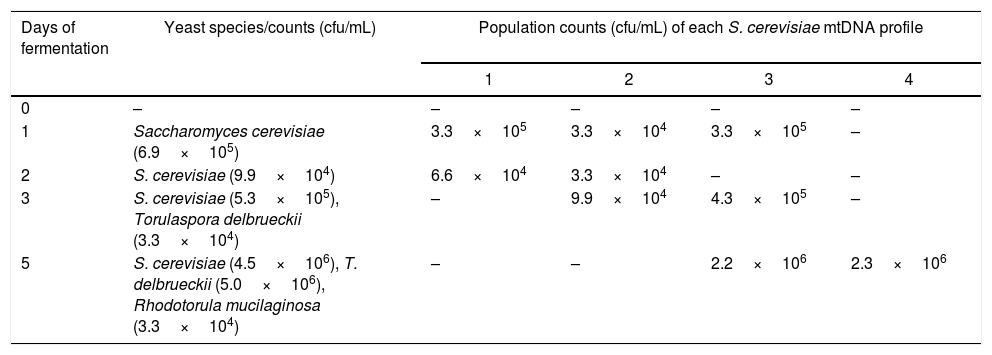
One sample of chicha de jora was studied during five days of fermentation (Table 2). At the start of fermentation, S. cerevisiae was present at a concentration of 6.6×105cfu/mL, and this species remained in the beverage until the fifth day, at which time counts were 4.5×106cfu/mL. Other yeast species such as T. delbrueckii and Rhodotorula mucilaginosa appeared after the third day of fermentation.
Mitochondrial restriction (mtDNA) profiles of the Saccharomyces cerevisiae strains isolated from chicha de jora during 5 days of fermentation and identified yeast species at each time point.
| Days of fermentation | Yeast species/counts (cfu/mL) | Population counts (cfu/mL) of each S. cerevisiae mtDNA profile | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| 0 | – | – | – | – | – |
| 1 | Saccharomyces cerevisiae (6.9×105) | 3.3×105 | 3.3×104 | 3.3×105 | – |
| 2 | S. cerevisiae (9.9×104) | 6.6×104 | 3.3×104 | – | – |
| 3 | S. cerevisiae (5.3×105), Torulaspora delbrueckii (3.3×104) | – | 9.9×104 | 4.3×105 | – |
| 5 | S. cerevisiae (4.5×106), T. delbrueckii (5.0×106), Rhodotorula mucilaginosa (3.3×104) | – | – | 2.2×106 | 2.3×106 |
Analysis of S. cerevisiae populations by restriction mtDNA profiles.
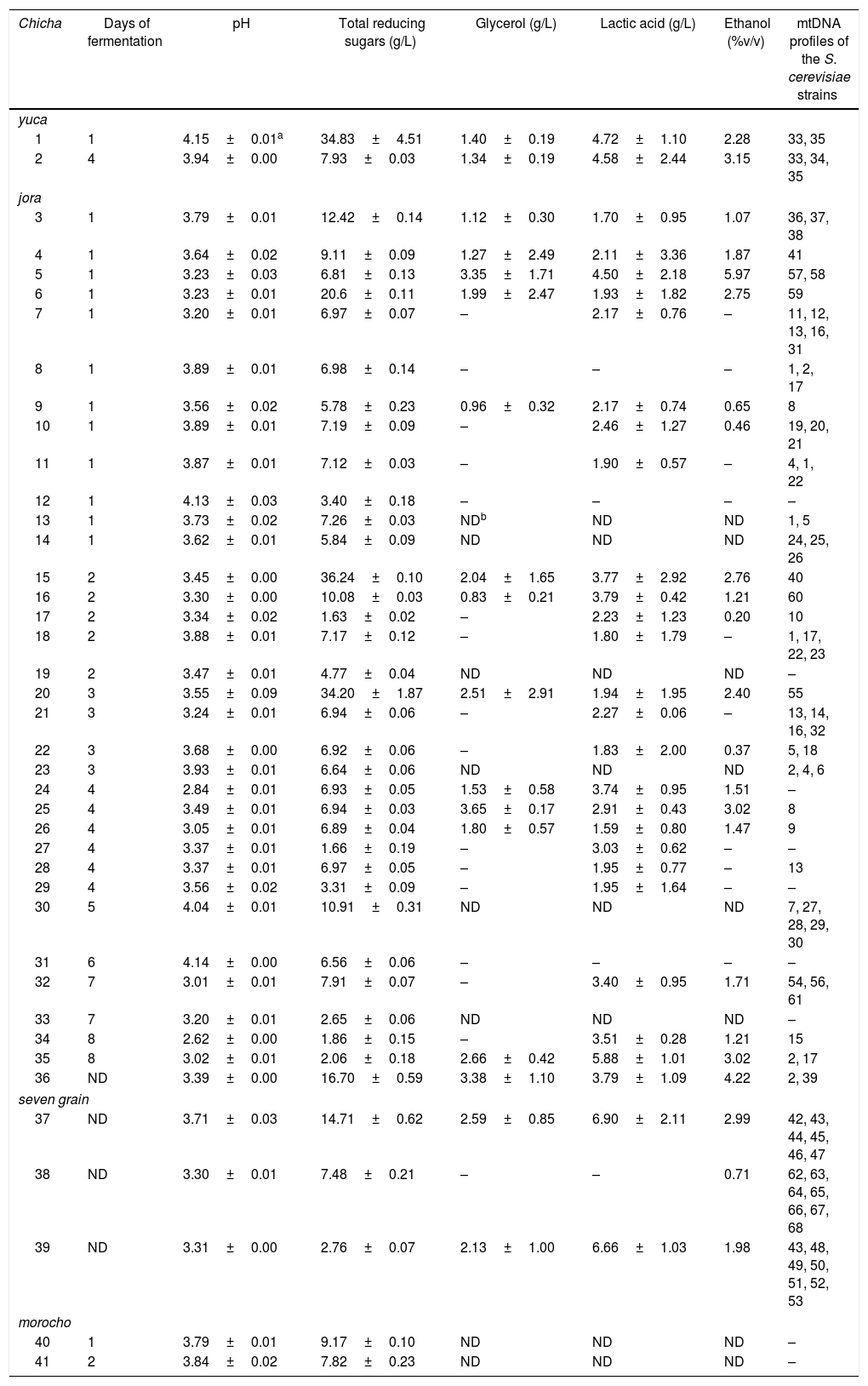
The 121 isolates of S. cerevisiae studied represented 68 different restriction mtDNA patterns. For chicha de yuca, 11 isolates of S. cerevisiae were analysed representing three different mtDNA restriction profiles in two samples studied. For seven-grain chicha, 28 isolates of S. cerevisiae were studied, representing 19 different mtDNA restriction profiles in three samples studied. In chicha de jora samples, 82 isolates of S. cerevisiae were analysed, and 46 mtDNA restriction profiles were found (Table 3). Two strains (mtDNA profiles 1 and 2) were found in four different chicha de jora samples, but most chicha samples yielded a set of strains with unique mtDNA restriction profiles that not occurs in another sample.
Physicochemical parameters of the chicha samples and the occurrence of the different mtDNA profiles of Saccharomyces cerevisiae strains in each sample.
| Chicha | Days of fermentation | pH | Total reducing sugars (g/L) | Glycerol (g/L) | Lactic acid (g/L) | Ethanol (%v/v) | mtDNA profiles of the S. cerevisiae strains |
|---|---|---|---|---|---|---|---|
| yuca | |||||||
| 1 | 1 | 4.15±0.01a | 34.83±4.51 | 1.40±0.19 | 4.72±1.10 | 2.28 | 33, 35 |
| 2 | 4 | 3.94±0.00 | 7.93±0.03 | 1.34±0.19 | 4.58±2.44 | 3.15 | 33, 34, 35 |
| jora | |||||||
| 3 | 1 | 3.79±0.01 | 12.42±0.14 | 1.12±0.30 | 1.70±0.95 | 1.07 | 36, 37, 38 |
| 4 | 1 | 3.64±0.02 | 9.11±0.09 | 1.27±2.49 | 2.11±3.36 | 1.87 | 41 |
| 5 | 1 | 3.23±0.03 | 6.81±0.13 | 3.35±1.71 | 4.50±2.18 | 5.97 | 57, 58 |
| 6 | 1 | 3.23±0.01 | 20.6±0.11 | 1.99±2.47 | 1.93±1.82 | 2.75 | 59 |
| 7 | 1 | 3.20±0.01 | 6.97±0.07 | – | 2.17±0.76 | – | 11, 12, 13, 16, 31 |
| 8 | 1 | 3.89±0.01 | 6.98±0.14 | – | – | – | 1, 2, 17 |
| 9 | 1 | 3.56±0.02 | 5.78±0.23 | 0.96±0.32 | 2.17±0.74 | 0.65 | 8 |
| 10 | 1 | 3.89±0.01 | 7.19±0.09 | – | 2.46±1.27 | 0.46 | 19, 20, 21 |
| 11 | 1 | 3.87±0.01 | 7.12±0.03 | – | 1.90±0.57 | – | 4, 1, 22 |
| 12 | 1 | 4.13±0.03 | 3.40±0.18 | – | – | – | – |
| 13 | 1 | 3.73±0.02 | 7.26±0.03 | NDb | ND | ND | 1, 5 |
| 14 | 1 | 3.62±0.01 | 5.84±0.09 | ND | ND | ND | 24, 25, 26 |
| 15 | 2 | 3.45±0.00 | 36.24±0.10 | 2.04±1.65 | 3.77±2.92 | 2.76 | 40 |
| 16 | 2 | 3.30±0.00 | 10.08±0.03 | 0.83±0.21 | 3.79±0.42 | 1.21 | 60 |
| 17 | 2 | 3.34±0.02 | 1.63±0.02 | – | 2.23±1.23 | 0.20 | 10 |
| 18 | 2 | 3.88±0.01 | 7.17±0.12 | – | 1.80±1.79 | – | 1, 17, 22, 23 |
| 19 | 2 | 3.47±0.01 | 4.77±0.04 | ND | ND | ND | – |
| 20 | 3 | 3.55±0.09 | 34.20±1.87 | 2.51±2.91 | 1.94±1.95 | 2.40 | 55 |
| 21 | 3 | 3.24±0.01 | 6.94±0.06 | – | 2.27±0.06 | – | 13, 14, 16, 32 |
| 22 | 3 | 3.68±0.00 | 6.92±0.06 | – | 1.83±2.00 | 0.37 | 5, 18 |
| 23 | 3 | 3.93±0.01 | 6.64±0.06 | ND | ND | ND | 2, 4, 6 |
| 24 | 4 | 2.84±0.01 | 6.93±0.05 | 1.53±0.58 | 3.74±0.95 | 1.51 | – |
| 25 | 4 | 3.49±0.01 | 6.94±0.03 | 3.65±0.17 | 2.91±0.43 | 3.02 | 8 |
| 26 | 4 | 3.05±0.01 | 6.89±0.04 | 1.80±0.57 | 1.59±0.80 | 1.47 | 9 |
| 27 | 4 | 3.37±0.01 | 1.66±0.19 | – | 3.03±0.62 | – | – |
| 28 | 4 | 3.37±0.01 | 6.97±0.05 | – | 1.95±0.77 | – | 13 |
| 29 | 4 | 3.56±0.02 | 3.31±0.09 | – | 1.95±1.64 | – | – |
| 30 | 5 | 4.04±0.01 | 10.91±0.31 | ND | ND | ND | 7, 27, 28, 29, 30 |
| 31 | 6 | 4.14±0.00 | 6.56±0.06 | – | – | – | – |
| 32 | 7 | 3.01±0.01 | 7.91±0.07 | – | 3.40±0.95 | 1.71 | 54, 56, 61 |
| 33 | 7 | 3.20±0.01 | 2.65±0.06 | ND | ND | ND | – |
| 34 | 8 | 2.62±0.00 | 1.86±0.15 | – | 3.51±0.28 | 1.21 | 15 |
| 35 | 8 | 3.02±0.01 | 2.06±0.18 | 2.66±0.42 | 5.88±1.01 | 3.02 | 2, 17 |
| 36 | ND | 3.39±0.00 | 16.70±0.59 | 3.38±1.10 | 3.79±1.09 | 4.22 | 2, 39 |
| seven grain | |||||||
| 37 | ND | 3.71±0.03 | 14.71±0.62 | 2.59±0.85 | 6.90±2.11 | 2.99 | 42, 43, 44, 45, 46, 47 |
| 38 | ND | 3.30±0.01 | 7.48±0.21 | – | – | 0.71 | 62, 63, 64, 65, 66, 67, 68 |
| 39 | ND | 3.31±0.00 | 2.76±0.07 | 2.13±1.00 | 6.66±1.03 | 1.98 | 43, 48, 49, 50, 51, 52, 53 |
| morocho | |||||||
| 40 | 1 | 3.79±0.01 | 9.17±0.10 | ND | ND | ND | – |
| 41 | 2 | 3.84±0.02 | 7.82±0.23 | ND | ND | ND | – |
Four different mtDNA profiles of S. cerevisiae were identified in chicha de jora studied during the five days of fermentation (Fig. 2).
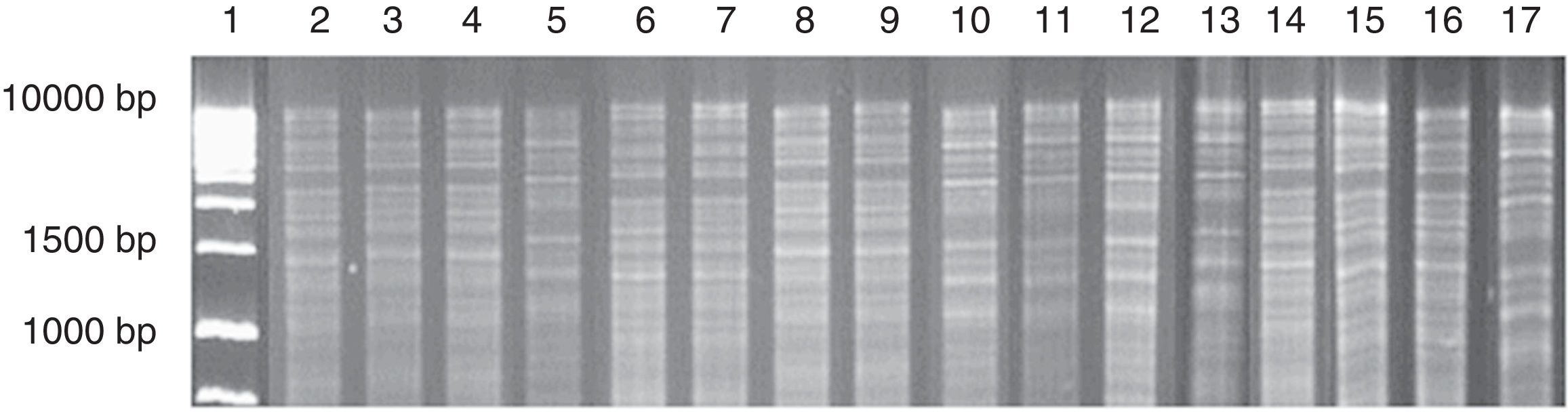
Mitochondrial DNA restriction patterns obtained from a chicha de jora sample at different times of fermentation. Column: 1: 1kb DNA ladder; 2–7: profiles found with 1 day of fermentation (2–4: pattern 1; 5: pattern 2; 6–7: pattern 3); 8–10: profiles found with 2 days of fermentation (8–9: pattern 1; 10: pattern 2); 11–15: profiles found with 3 days of fermentation (11–13: pattern 2; 14–15: pattern 3); 16–17: profiles found with 5 days of fermentation (16: pattern 3; 17: pattern 4).
Only the S. cerevisiae strain with mtDNA profile 3 occurred from the first to the fifth day of fermentation (Table 2). Strains with mtDNA profiles 2 and 3 showed increasing counts over the course of the fermentation. In contrast, the strain with mtDNA profile 1 decreased in number during the fermentation process, and after three days it was not detected.
Physicochemical analysesThe highest pH value was 4.15, in sample 1 after 24h of fermentation, and the lowest value was 2.62, in sample 35 after 8 days of fermentation (Table 3). The lowest value of total reducing sugars was 1.63g/L and the highest value was 36.24g/L, both found in chicha de jora samples. The largest amount of glycerol was 3.65g/L, found in chicha de jora. Lactic acid contents ranged between 1.59 and 6.90g/L. Ethanol was not detected in 10 chicha samples. The highest value of ethanol was 5.97 (%v/v) in sample 5 of chicha de jora after 24h of fermentation.
DiscussionOur results showed that in most chicha samples studied, S. cerevisiae was the predominant species, followed by T. delbrueckii, P. kudriavzevii, C. sake, D. bruxellensis, P. fermentans, and S. ludwigii, but generally at much lower counts. All these yeast species may contribute to the sensory quality of the beverage. S. cerevisiae was also the dominant species in two productions a maize chicha prepared by local producers from Maimará and Tumbaya villages (Quebrada de Humahuaca region) in Northwest of Argentina.7 However, Candida parapsilosis and an undescribed Pichia species were the second and third most abundant yeasts in these fermentations in Argentina. Vallero et al.5 isolated only S. cerevisiae from samples of chicha de jora in 10 traditional “chicherías” in Cusco region in Peru. These results show that the fermentation process associated with maize-based chicha is carried by S. cerevisiae and other non-Saccharomyces species that occur in minor frequencies. The non-Saccharomyces species found in our study can be isolated from different substrates worldwide, including those of fermentation processes for beverage production.1,5,20–22 The origin of the non-Saccharomyces species associated with the chicha samples studied might be explained by the different manufacturing processes for these beverages. Chicha de jora is prepared in different ways, employing a wide variety of raw materials with additional ingredients such as fruits, herbs, spices, brown sugar, and sugar. Seven-grain chicha is produced with seven varieties of maize flour and chicha de yuca with few raw materials including cassava and the seed of the Ungurahua palm. Chicha de morocho, despite also being manufactured from a wide variety of ingredients, was collected at restaurants in La Mariscal, a district of Quito (data not shown), where the beverage was produced following Good Manufacturing Practice. Some ingredients used in the chicha manufacturing process, as fruits, herbs and spices, are added after boiling the jora flour. Based on this information, the yeast populations could originate from these ingredients used in beverage preparation, and from other sources such as handlers, utensils, and vessels used during the fermentation process.
A high number of different mtDNA profiles of S. cerevisiae were found associated with the chicha production process in our study. The occurrence of these different mtDNA profiles of S. cerevisiae associated with traditional beverages is well documented.21,23,24 In addition, the occurrence of different strains during the fermentation process, as observed in the chicha de jora studied during the five days of fermentation is very common in spontaneous fermentation during traditional beverage production.21,25,26 As each mtDNA profile represents a different genetic strain of S. cerevisiae, these strains could be contributing to different sensory proprieties of the beverage. The selection of the best S. cerevisiae strains for chicha production is interesting, and these studies could be based on strain diversity with different mtDNA restriction profiles occurring during the fermentative process.
The low pH values of the chicha studied might be associated with acid content, specifically, lactic and acetic acids, produced during fermentation. These organic acids can contribute negatively to the sensory quality of the beverage, as off-flavours can result when these acids are present in high amounts.23 The total reducing sugars varied widely, possibly due to sampling at different fermentation times and the diversity of raw materials used in chicha preparation. This variation might be related to the efficiency of the microbial community in utilizing these reducing sugars. Glycerol is an alcohol of great importance in alcoholic beverages, as it provides a sweet aroma and contributes to the viscosity of the final product. The largest amount of glycerol was 3.65g/L, found in chicha de jora. It is important to note that glycerol in high amounts is not desirable because it gives a stale flavour to the beverage.27
Altay et al.28 noted physicochemical parameters very similar to those found in this study when studying shalgam juice, a fermented non-alcoholic beverage produced from the lactic fermentation of black carrot. According to the authors, the pH of this beverage was between 3.15 and 4.25 and the major fermentation products obtained were lactic acid (from 5.18 to 8.05g/L), acetic acid (0.57–0.83g/L), ethanol (0.79–6.41%) and volatile aromatic compounds. Low concentrations of ethanol present in some samples and the presence of lactic acid make the possibility that fermentation by lactic acid bacteria instead of alcoholic fermentation had occurred in major scale in these samples. With respect to boza, a beverage fermented by yeasts and lactic acid bacteria, the pH ranged from 3.16 to 4.63 and the ethanol content ranged from not detectable to 0.39%. Mendoza et al.7 reported ethanol concentration around 1% in Argentinean maize-based chichas, result similar to the chichas analysed in our study. Therefore, the chicha samples studied presented generally similar physicochemical parameters to those observed for other traditional fermented beverages. Finally, the Ecuadorian chichas can be considered acid fermented beverages, and the yeast species associated with its fermentative process should contribute to the singular aroma and unique flavour of this beverage.
Conflicts of interestThe authors declare no conflicts of interest.
This work was funded by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq – Brazil), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG – Brazil), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES – Brazil), and the Secretaría Nacional de Educación Superior, Ciencia, Tecnología e Innovación (Ecuador).