Shigatoxigenic and enteropathogenic Escherichia coli with virulence and multidrug resistance profile were isolated from Nile tilapia. This study finding is of great importance to public health because they help understand this pathogen epidemiology in fish and demonstrate how these animals can transmit E. coli related diseases to humans.
Escherichia coli (E. coli) is not a natural inhabitant of the fish microbiota, nevertheless, it can be isolated from these animals gut due to its presence in contaminated aquatic environments.1 It is worth noticing that this microorganism have pathogenic strains standing out as emerging zoonotic potential, as well as shigatoxigenic (STEC) and enteropathogenic (EPEC) E. coli. STEC strains produce the shiga toxin (Stx), which is its main virulence factor. There are two classes of shiga toxin, Stx1 and Stx2, with the last one presenting seven subtypes.2 The EPEC may be either typical or atypical, with the atypical strains do not carrying virulence factor that encodes the bundle-forming pilus (bfp), but it carries the eae gene, that is located at the locus of enterocyte effacement (LEE), which is a pathogenicity island, that promote attaching and effacing lesions (A/E). The ability to induce A/E lesions is mediated by genes located on the LEE, as well as additional ones that are outside of it.3
Several studies have analyzed STEC and EPEC, and their virulence in humans,4 cattle,5 sheep,6 pigs,7 and buffaloes.8 However, only a few studies have analyzed the presence of STEC and EPEC in fish9,10 and, of these, none has detected presence of adhesion and ESBL genes. In addition, none has performed the stx2 subtyping in STEC strains from fish. In this regard, this pioneer study aimed to compare the prevalence of STEC and EPEC strains in intensively farmed fish and free-living fish; as well as to detect their virulence and antibiotic resistant profile and analyze their genetic similarity looking for how these fishes contribute to humans infections.
The Ethics Committee on Animal Use (CEUA) approved this study under the protocol number 04076/14. Primers used are described in Table 1. The samples were collected from the fish species Oreochromis niloticus, from six different fish farms and three ranches located at northeast region of Sao Paulo state. A total of 472 samples were collected. Three hundred and seventy three (373) samples were obtained from fish farm animals and of these, 275 were from stools, 80 from muscles and 18 from the nurseries water. The other 99 samples were obtained from free-living fish, these been 90 from stools and nine from the river water. Samples were transferred to tubes containing BHI broth (Brain Heart Infusion) and after an incubation period, the DNA were extracted by thermal lysis according to Borges.7
Information about the primers used in PCR reactions.
| Primers | Sequences | Size | Reference |
|---|---|---|---|
| stx1-F | AGAGCGATGTTACGGTTTG | 388 | 32 |
| stx1-R | TTGCCCCCAGAGTGGATG | ||
| stx2-F | TGGGTTTTTCTTCGGTATC | 807 | 32 |
| stx2-R | GACATTCTGGTTGACTCTCTT | ||
| eae -F | AGGCTTCGTCACAGTTG | 570 | 32 |
| eae -R | CCATCGTCACCAGAGGA | ||
| bfpA - F | GGAAGTCAAATTCATGGGGGTAT | 300 | 33 |
| bfpA -R | GGAATCAGACGCAGACTGGTAGT | ||
| ehxA -F | GGTGCAGCAGAAAAAGTTGTA G | 340 | 34 |
| ehxA -R | TCTCGCCTGATAGTGTTTGGT A | ||
| saa- F | CGTGATGAACAGGCTATTGC | 119 | 17 |
| saa -R | ATGGACATGCCTGTGGCAAC | ||
| iha -F | CAGTTCAGTTTCGCATTCACC | 1305 | 35 |
| iha -R | GTATGGCTCTGATGCGATG | ||
| toxB -F | ATACCTACCTGCTCTGGATTGA | 602 | 36 |
| toxB -R | TTCTTACCTGATCTGATGCAGC | ||
| efa1- F | GAGACTGCCAGAGAAAG | 479 | 37 |
| efa1-R | GGTATTGTTGCATGTTCAG | ||
| lpfAO113- F | ATGAAGCGTAATATTATAG | 573 | 38 |
| lpfAO113- R | TTATTTCTTATATTCGAC | ||
| lpfAO157/OI-141-F | CTGCGCATTGCCGTAAC | 412 | 39 |
| lpfAO157/OI-141-R | ATTTACAGGCGAGATCGTG | ||
| lpfAO157/OI-154-F | GCAGGTCACCTACAGGCGGC | 525 | 34 |
| lpfAO157/OI-154- R | CTGCGAGTCGGCGTTAGCTG | ||
| astA - F | TCGGATGCCATCAACACAGT | 125 | 24 |
| astA-R | GTCGCGAGTGACGGCTTTGTAG | ||
| paa -F | ATGAGGAACATAATGGCAGG | 360 | 23 |
| paa-R | TCTGGTCAGGTCGTCAATAC | ||
| CTX-M group 1- F | TTAGGAARTGTGCCGCTGYA | 668 | 11 |
| CTX-M group 1- R | CGATATCGTTGGTGGTRCCAT | ||
| CTX-M group 2 - F | CGTTAACGGCACGATGAC | 404 | 11 |
| CTX-M group 2 - R | CGATATCGTTGGTGGTRCCAT | ||
| CTX-M group 8/25 F | AACRCRCAGACGCTCTAC | 326 | 11 |
| CTX-M group 8/25R | TCGAGCCGGAASGTGTYAT | ||
| CTX-M group 9 - F | TCAAGCCTGCCGATCTGGT | 561 | 11 |
| CTX-M group 9 - R | TGATTCTCGCCGCTGAAG | ||
| TEM - F | CATTTCCGTGTCGCCCTTATTC | 800 | 11 |
| TEM - R | CATTTCCGTGTCGCCCTTATTC | ||
| SHV - F | AGCCGCTTGAGCAAATTAAAC | 713 | 11 |
| SHV - R | ATCCCGCAGATAAATCACCAC | ||
| stx2a-F | GCGATACTGRGBACTGTGGCC | 347 | 2 |
| stx2a-R | GCCACCTTCACTGTGAATGTG | ||
| stx2b-F | AAATATGAAGAAGATATTTGTAGCGGC | 251 | 2 |
| stx2b-R | CAGCAAATCCTGAACCTGACG | ||
| stx2c-F | GAAAGTCACAGTTTTTATATACAACGGGTA | 177 | 2 |
| stx2c-R | CCGGCCACYTTTACTGTGAATGTA | ||
| stx2d-F | AAARTCACAGTCTTTATATACAACGGGTG | 179 | 2 |
| stx2d-R | TTYCCGGCCACTTTTACTGTG | ||
| stx2e-F | CGGAGTTACGGGGAGAGGC | 411 | 2 |
| stx2e-R | CTTCCTGACACCTTCACAGTAAAGGT | ||
| stx2f-F | TGGGCGTCATTCACTGGTTG | 424 | 2 |
| stx2f-R | TAATGGCCGCCCTGTCTCC | ||
| stx2g-F | CACCGGGTAGTTATATTTCTGTGGATATC | 573 | 2 |
| stx2g-R | GATGGCAATTCAGAATAACCGCT | ||
| adk-F | ATTCTGCTTGGCGCTCCGGG | 583 | a |
| adk-R | CCGTCAACTTTCGCGTATTT | ||
| fumC-F | TCACAGGTCGCCAGCGCTTC | 806 | a |
| fumC-R | GTACGCAGCGAAAAAGATTC | ||
| gyrB-F | TCGGCGACACGGATGACGGC | 911 | a |
| gyrB-R | ATCAGGCCTTCACGCGCATC | ||
| icd-F | ATGGAAAGTAAAGTAGTTGTTCCGGCACA | 878 | a |
| icd-R | GGACGCAGCAGGATCTGTT | ||
| mdh-F | ATGAAAGTCGCAGTCCTCGGCGCTGCTGGCGG | 932 | a |
| mdh-R | TTAACGAACTCCTGCCCCAGAGCGATATCTTTCTT | ||
| purA-F | CGCGCTGATGAAAGAGATGA | 816 | a |
| purA-R | CATACGGTAAGCCACGCAGA | ||
| recA | CGCATTCGCTTTACCCTGACC | 780 | a |
| recA | AGCGTGAAGGTAAAACCTGTG |
Screening for the detection of STEC and EPEC strain were based on the, stx1, stx2 and eae genes detection by multiplex PCR.7 When one of these genes were detected, individual colonies from each sample were tested by PCR to isolate STEC and EPEC strains according to the protocol available at www.apzec.ca/en/APZEC/Protocols/pdfs/ECL_PCR_Protocol.pdf. This methodology is in accordance to the OIE Reference Laboratory for Escherichia coli (EcL – Faculté de Médecine Véterinaire, Université de Montréal). Isolates were further submitted to another PCR to detect others virulence genes as follow: bfpA, ehxA, saa, iha, toxB, efa1, lpfAO113, lpfAO157/OI-141, lpfAO157/OI-154, astA and paa genes. The Stx2 variants analysis was performed by stx2 subtyping according to Scheutz.2
The antimicrobial susceptibility test was performed using the disc diffusion method.30 The antimicrobials chosen were the ones most used in fish farming and which are important for the detection of resistance genes dissemination. In this regard, drugs tested were ampicillin (10μg), cephalothin (30μg), streptomycin (10μg), gentamicin (10μg), ciprofloxacin (5μg), chloramphenicol (30μg), tetracycline (30μg), nitrofurantoin (300μg), nalidixic acid (30μg), sulfamethoxazole and trimethoprim (25μg), ceftriaxone (30μg), cefoxitin (30μg), kanamycin (30μg), norfloxacin (10μg), enrofloxacin (5μg) (Oxoid). In addition, E. coli isolates were screened for extended-spectrum beta-lactamase (ESBL) genes for the blaCTX-M genotype groups 1, 2, 8, 9 and 25, the blaTEM, and the blaSHV.11
Phylogenetic E. coli groups’ classification was performed according to the methodology proposed by Clermont.12 Serotyping was performed at the E. coli Reference Center (EcRc) at Pennsylvania State University. The O somatic antigen were determinate by agglutination plates, also the PCR-RFLP of fliC gene, which encodes flagella, were performed to determine the H flagella antigen. Somatic antigens used were O1 to O187, with the exception of O31, O47, O67, O72, O94, O122 and the flagellar antigens used were H1 to H49, except H17, since these serogroups still not have been designated.
The isolates were also characterized by PFGE pattern of the PulseNet protocol as described by Ribot.13 Briefly, the chromosomal DNA was digested with Xba1 and the electrophoresis conditions were an initial time of 2.2s and an end time of 54.2s in a gradient of 6V and the gels were electrophoresed for 21h. The fragment similarities were compared using the Dice coefficient and the dendrogram was constructed by neighbor-joining grouping using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). MLST was performed following the Achtmans's scheme (http://mlst.ucc.ie/mlst/dbs/Ecoli), through the sequencing of the PCR amplification products of the adk, fumC, gyrB, icd, mdh, purA, and recA genes. The generated sequences were trimmed and analyzed by the Phred/Phrap/Consed software package.
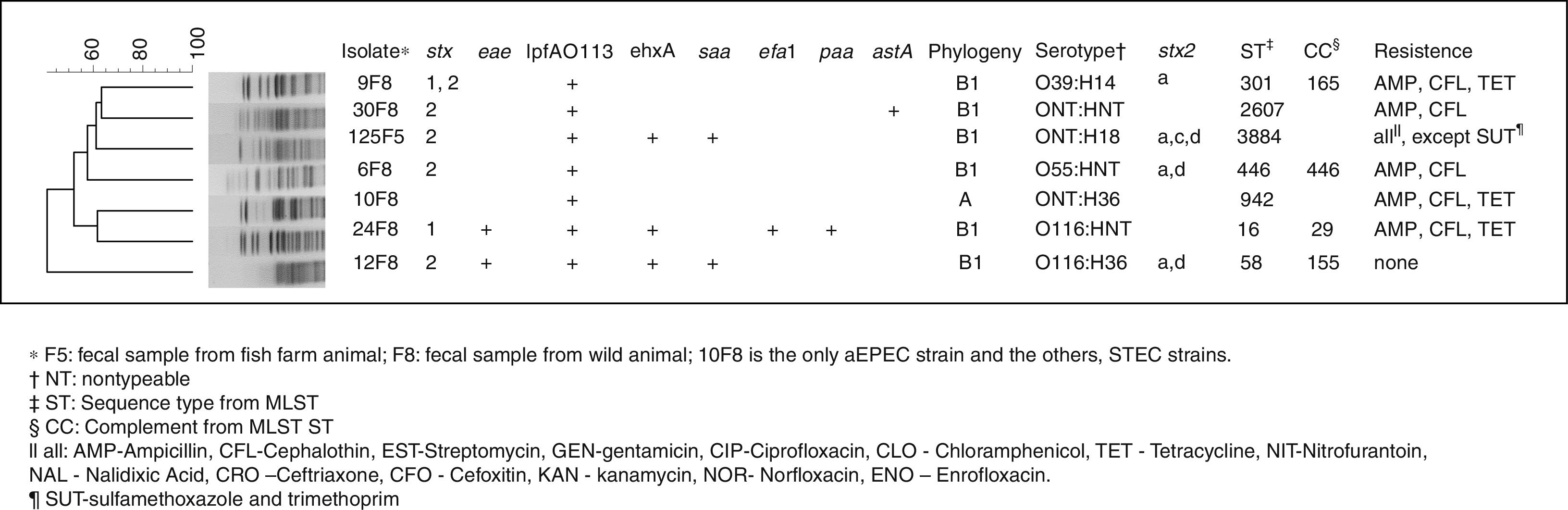
All the results are shown in Fig. 1. Of the 373 analyzed samples from the fish farm, one (0.2%), from stools, tested positive for a STEC related gene (isolate 125F5). Of the 99 free-living fish analyzed samples, six (6%), also from stools, were positive for at least one of the STEC or EPEC related genes (isolates 6F8, 9F8, 10F8, 12F8, 24F8 and 30F8). In addition, all six isolates were collected from the same location, and the stx1, stx2 and eae genes were detected. None of the muscle or water samples tested were positive for the STEC or EPEC markers investigated. Isolates from the fish farms were positive for ehxA, lpfAO113 and saa virulence genes. Also, strains from the free-living fish presented astA, ehxA, lpfAO113, saa, efa1 and paa genes. Regarding Stx2 toxin variants, the subtypes stx2a, stx2c and stx2d were observed at the same isolate.
From the 15 antimicrobial drugs tested, the isolates originated from the fish farm animals were resistant to 14 of them, while the isolates from the free-living fish were resistant to three antimicrobials and no ESBL genes were found. All of the STEC strains belong to group B1 and the aEPEC strain to group A as in accord with the classification of Clermont.12 From seven isolates analyzed by serotyping, three were nontypeable for the O antigen, and three isolates were nontypeable for the H antigen. Thus, the groups detected were O55, O39, O116, H14, H18 and H36; and their serotype is shown in Fig. 1. Seven isolates possessed a heterogeneous profile, by PFGE analysis, and seven distinct sequence types (STs) with four clonal groups (CCs) detected by the MLST technique.
Although STEC and aEPEC strains isolated from fish are not natural inhabitants of its microbiota, these strains can colonize the fish through a contaminated environment of which they live.1 In both establishments that these positive strains were isolated, presence of cattle were observed around the nurseries and rivers. It is important to notice that bovine is considered the main reservoir of pathogenic E. coli.26
Moreover, in this study, all muscle and water samples were negative for the presence of STEC or EPEC. This result should not be taken lightly, because the samples were collected by dissecting the animal using aseptic conditions so that muscles samples were carefully separated from the intestinal content, which it does not occur at the fishermen or slaughterhouses daily practice. Commonly, a cut is made between the anus and the fish’ head, releasing all of its intestinal contents and, in the process, contaminating the muscle. In this regard, according to Kim,31 pathogenic E. coli can enter the human food chain mainly through food contamination. Moreover, none of the water sample tested positive for STEC or EPEC and this was associated with the large water flow at the nurseries and rivers.
These pathogens have already detected in Brazil4,6–8,14; and in other countries such as United States,15 Argentina,16 Netherlands,26 Iran,18 Tunisia,19 and Australia.20 These studies, as well as the present one, are fundamental to understand these pathogens epidemiology, since they have great importance in animal and public health.
In this study, one STEC strain with eae gene also contained several other genes, efa1, ehxA, lpfAO113 and paa, which have been associated with cases of diarrhea.21,22 The presence of saa gene was shown to be closely related to the presence of the ehxA gene in STEC strains devoid of eae gene, regardless of their serotype.4,23 In an STEC isolated, the presence of astA gene, which is important for pathogenesis of diarrhea and plays a key role in this strain’ virulence,24 was observed. Also, the lpfAO113 gene was shown in previous reports to have a high prevalence in STEC isolated from different animals.4 Regarding Stx2 toxin variants, the stx2a, stx2c and stx2d subtypes were observed in the same isolate; this combination is very unusual and makes this strain fairly unique and with aggravated virulence. The presence of stx2a, stx2c and stx2d subtype are often associated with the hemorrhagic colitis and with hemolytic uremic syndrome.2
The quinolones, tetracyclines, aminoglycosides and amphenicols are the most commonly used antimicrobials in fish farming25 and thus, it may explain the multiresistance observed in the fish farm isolates. All of the tested quinolones, tetracyclines, aminoglycosides and amphenicols were ineffective. Although it was observed multiresistance, no ESBL genes were found. The ESBL would confer an even greater risk for the raw fish consuming population, because these bacteria's can produce an enzyme that are able to hydrolyze the beta lactam ring of penicillins, cephalosporins and aztreonam, thus conferring resistance to these antimicrobials.11 However, this multiresistance profile shows a phenotypic response from the isolates, and due to the selective pressure originated from the abusive use of broad-spectrum cephalosporins, according to data obtained in the present study, ESBL strains can emerge rapidly, as it is already observed in other animal productions.
According to the phylogenetic results of this study, it is confirmed that tropical populations can harbor strains of group A and B1 preferentially, which may be one of the factors that could explain tropical countries higher diarrhea frequency.27 Although much is reported about the O157 strains, non-O157 STEC strains are the most prevalent in animals and food. For this reason, chances of humans becoming infected by these strains are large, indicating the importance of this serogroups to public health.5 Therefore, the O116 serogroup observed in the present study is relevant due to the fact that it is often associated with severe human disease. Furthermore, the strain with the flagelar antigen H18 which contains the stx2 but not the eae gene, has also shown frequent association with infections in animals.28 Moreover, the ONT: H18 serotype in eae negative and saa positive strains, similarly to the ones in this study, has previously been detected in pathogenic E. coli in cattle,29 thus emphasizing the fact that these animals were, likely, the source of this pathogen infection in fish.
The genetic diversity analysis showed that, although the isolates belonged to the same bacterial species, they had genetic diversities that were highlighted though the PFGE technique. The same was observed with the MSLT data, indicating that although they had a common ancestral origin, the transference of genetic information, though time, made this isolates very diverse, thus explaining their distinct phylogenetic classification.
Ours results shows that fish can harbor an important combination of Stx2 subtypes and putative adhesions genes. Also, it draws attention to the fact that the indiscriminate use of antibiotics in fish farming has the potential to endanger consumer health through the dissemination of antibiotic resistance genes. And finally, it highlights the role of STEC and aEPEC as foodborne pathogens in fish for human consumption.
The authors would like to thank FAPESP for all research support granted (2011/07358-2 and 2011/15050-8).