Discharge of coke-oven wastewater to the environment may cause severe contamination to it and also threaten the flora and fauna, including human beings. Hence before dumping it is necessary to treat this dangerous effluent in order to minimize the damage to the environment. Conventional technologies have inherent drawbacks however, biological treatment is an advantageous alternative method. In the present study, bacteria were isolated from the soil collected from the sites contaminated by coke-oven effluent rich in phenol and cyanide. Nucleotides sequence alignment and phylogenetic analysis showed the identity of the selected phenol and cyanide degrading isolates NAUN-16 and NAUN-1B as Pseudomonas putida and Pseudomonas stutzeri, respectively. These two isolates tolerated phenol up to 1800mgL−1 and cyanide up to 340mgL−1 concentrations. The isolates were immobilized on activated charcoal, saw dust and fly ash. The effluent was passed through the column packed with immobilized cells with a flow rate of 5mLmin−1. The isolates showed degradation of phenol up to 80.5% and cyanide up to 80.6% and also had the ability to reduce biological oxygen demand, chemical oxygen demand and lower the pH of effluent from alkaline to near neutral. The study suggests the utilization of such potential bacterial strains in treating industrial effluent containing phenol and cyanide, before being thrown in any ecosystem.
A wide variety of synthetic chemicals have found their way into the ecosystem as a consequence of industrial activities, agricultural applications and use in domestic purposes. The rate of discharge of these materials into the ecosystem is more than that of their degradation by natural processes, resulting in the accumulation of such substances in the environment.1 The toxic pollutants phenol and cyanide are amongst the most common, potentially dangerous substances that are appearing in nature due to industrialization.2 Coke-oven based industries and steel plants release phenol and cyanide simultaneously (in the effluent discharged) thus causing a threat to the ecosystem. Coke-oven process wastewater contains phenol (150–2000mgL−1) and cyanide (0.1–0.6kgton−1 of coke) at high levels far exceeding the normal limits.3
Phenol present in effluent from coke-oven units is a hazardous pollutant, which is poisonous to animals and toxic to soil microbes and some aquatic species at concentrations in the low mgL−1 range.4,5 Furthermore, phenol has an impact on human health and it is recommended that human exposure should not exceed 20mg on an average working day.6 Phenol is placed in the list of priority contaminants by the U.S. Environmental Protection Agency.7 The coke-oven effluent also contains an unacceptably high level of cyanide (a potent inhibitor of respiration due to its extreme toxicity towards cytochrome oxidase). Therefore, to protect the environment from such toxic materials, the wastewater and industrial effluents must be suitably treated before discharge for sustainable environment and agriculture.8,9
New technologies for waste disposal that use high-temperature incineration and chemical decomposition (e.g., base-catalyzed dechlorination, UV oxidation) have evolved. Although they can be very effective at reducing contaminants but at the same time have several drawbacks. These methods are complex, uneconomical and lack public acceptance because of the formation of toxic intermediates, cost ineffectiveness and associated hazards. The associated deficiencies in these methods have focused efforts toward harnessing modern-day bioremediation process as a suitable alternative.10,11 Bioremediation, especially associated with microorganisms, used for the cleanup of contaminated waters and soils is receiving most attention because of its environmental friendly approach and its ability to completely mineralize toxic organic compounds.12,13 It is also a cost-effective strategy.14,15 Besides cost-effectiveness, it is a permanent solution, which may lead to complete mineralization of the pollutant. Furthermore, it is a non-invasive technique, leaving the ecosystem intact.16
The purpose of the study was to find bacterial strains that can be used as alternatives to conventional technologies to treat coke-oven wastewater with the simultaneous biodegradation of phenol and cyanide present in coke-oven effluent.
Materials and methodsOrganism source and culture conditionsSoil samples were collected in pre sterilized bottles from contaminated site in district Haridwar, Uttarakhand, India (29.9°N and 78.17°E), where coke-oven effluent was being dumped since several years. For isolation of bacteria, one gram of soil suspension was serially diluted in sterile test tubes containing sterile distilled water. The contents were spread out on nutrient agar (NA) plates and incubated at 28°C for 24h.17 Colonies that grew on plates were selected for further studies.
Growth studies in presence of phenol and cyanideThe isolates were grown in nutrient broth (NB) medium and NB-containing phenol (Phenol GR, Merck) (0–2000mgL−1) and cyanide (KCN, Merck) (0–400mgL−1) in a batch culture. The bacteria were grown at 160rpm, incubation temperature 28°C and pH 7.0. After inoculation, the growth mass was measured as OD610 (UV-VIS Spectrophotometer Genesys 6) at 2h-intervals, up to 48h.18 The isolates were screened for further studies on the basis of their tolerance to phenol and cyanide.
Identification of bacteriaThe selected isolates were microscopically checked for their morphological characteristics. Their biochemical identities were elucidated by various tests according to Bergey's Manual of Systematic Bacteriology.19 Selected isolates (NAUN-16 and NAUN-1B) on the basis of tolerance towards phenol and cyanide were further identified by 16S rRNA gene sequencing. For this, bacterial DNA was isolated20 and evaluated on 1.2% agarose gel.21 For sequence analysis of ∼1.5kb 16S rRNA gene fragment, consensus universal forward and reverse primers fD1 and rP2 respectively were used and amplification performed by Taq DNA Polymerase. Reaction Mixture components for 50μL final volume is reaction buffer (10X): 5.0μL, dNTPs mix (10mM): 5.0μL, MgCl2 (25mM): 3.0μL, forward primer (10pmol/μL): 1.5μL, reverse primer (10pmol/μL): 1.5μL, template DNA: 5.0μL, Taq DNA polymerase (5U/μL): 0.5μL and MQ water: 28.5μL. The PCR product was bi-directionally sequenced using the forward and reverse primer under the conditions: Initial denaturation (95°C) for 2min, denaturation (95°C) for 1min, annealing (48°C) for 1min, elongation (72°C) for 1min, repetition from step 2 to 4 for 29 cycles and finally elongation (72°C) for 5min. The 16S rDNA gene sequence was used to carry out BLAST22 with the NCBI GenBank database.23 First ten sequences were aligned using Clustal W. Distance matrix was generated using RDP database24 and the phylogenetic tree was constructed in MEGA 4 software25 using Neighbor Joining method26 for identification of bacterial isolates.
Treatment of effluent by immobilized cellsActivated charcoal (Merck), saw dust (of Sesamum indicum obtained from local saw mill) and fly ash (collected from thermal power plant) were procured to use as carrier material for immobilization of bacterial cells.
To determine pH, 10g solid carrier was mixed with 90mL of distilled water with thorough stirring and pH of mixture was determined.27 The moisture content of each carrier was determined on a wet and dry weight basis. Water-holding capacity of each carrier was determined by adding 100g oven-dried carrier material in a 500mL size beaker containing appropriate amount of water for dipping the material. Saturated material was decanted on gauze to remove extra water and weighed.27
The bacterial suspension used for immobilization contained 12h grown cells of the screened bacteria incubated at 28°C in the nutrient broth. Broth cultures were added separately to each carrier in the ratio 1:3 and thoroughly mixed to obtain a homogeneous carrier-inoculum mixture. This mixture was spread in the trays and kept for 48h curing at room temperature.27 Carrier with un-inoculated broth served as control. Each inoculated and uninoculated carrier was filled up to 1/3 in 60cm glass columns (Borosil). The column diameter used was 7cm so as to allow the effluent run at an optimum flow rate of 5mLmin−1.28
Physico-chemical analysis of treated and untreated effluentThe physico-chemical experiments were conducted taking the untreated and treated effluent samples. Biological oxygen demand (BOD), chemical oxygen demand (COD) and pH levels were estimated before and after the treatment. Phenol in the effluent sample was estimated colorimetrically using 4-aminoantipyrine method. In this method phenol reacts with 4-aminoantipyrine in the presence of potassium ferricyanide to form a colored antipyrine dye. Chloroform was used to extract the dye from aqueous solution. The absorbance of the extract was read at 460nm using Genesys 6 spectrophotometer. Cyanide in the effluent sample was estimated by pyridine barbituric acid colorimetric method (575nm) after distillation. The procedures adopted for analyses were according to the American Public Health Association Manual.17
Survival of immobilized cellsPseudomonas putida (NAUN-16) and Pseudomonas stutzeri (NAUN-1B), selected for the study, were marked with antibiotic resistance genes so as to easily isolate and count them from mixed culture. P. putida NAUN-16strep+ was indigenously resistant to streptomycin (50mgL−1). The plasmid vector pBC16 was used to engineer tetracycline-resistance (200mgL−1) taking donor Bacillus cereus strain GP7 (pBC16), as described by Bernhard et al.29 While P. stutzeri NAUN-1Bamp+ indigenously resistant to ampicillin (50mgL−1) was engineered using Tn5 delivery suicide vector pGS9 to cause streptomycin-resistance (100mgL−1) taking donor Escherichia coli strain WA803 (pGS9), as described by Kumar et al.30 Subsequently the strains were designated P. putida NAUN-16strep+tet+ and P. stutzeri NAUN-1Bamp+strep+.
Carrier-inoculum mixtures were stored separately at room temperature (25±5°C) conditions. After every 30 days, the stored samples were analyzed to determine the number of viable cells up to six months (180 days). Population density was also monitored before and after the treatment process. The colony forming units (cfu) were measured taking 1g of carrier (with immobilized cells) from the columns which was run for 7 days. The carrier taken from the column was re-suspended in 10mL sterile normal saline and left to rehydrate for 2min. Subsequently, each suspension was agitated vigorously on a vortex mixture for 10min to suspend the adhered cells. Aliquots containing 0.1mL were plated on nutrient agar supplemented with streptomycin (50mgL−1) and tetracycline (200mgL−1) for P. putida NAUN-16strep+tet+ and with ampicillin (50mgL−1) and streptomycin (100mgL−1) for P. stutzeri NAUN-1Bamp+strep+ and incubated at 28°C for 48h before final counts were taken.31
Results and discussionPhenol and cyanide resistant isolatesMicroorganisms have the ability to adapt to a variety of environmental conditions. The microorganisms able to use the pollutants as a nutrient source can efficiently degrade the target compounds.32 In total 15 bacterial strains were isolated from the contaminated soil on NA medium. Two bacterial isolates designated NAUN-16 and NAUN-1B were selected on the basis of their growth in the presence of phenol and cyanide in vitro conditions. The isolate NAUN-16 was Gram negative, motile, rod, endospore-forming and catalase positive and glucose fermenting bacteria. Isolate NAUN-1B was Gram negative, rod, gelatinase, oxidase and catalase positive, and denitrifying bacteria. On the basis of 16S rRNA gene sequence (1.5kb) analysis isolates NAUN-16 and NAUN-1B were identified as P. putida and P. stutzeri respectively.
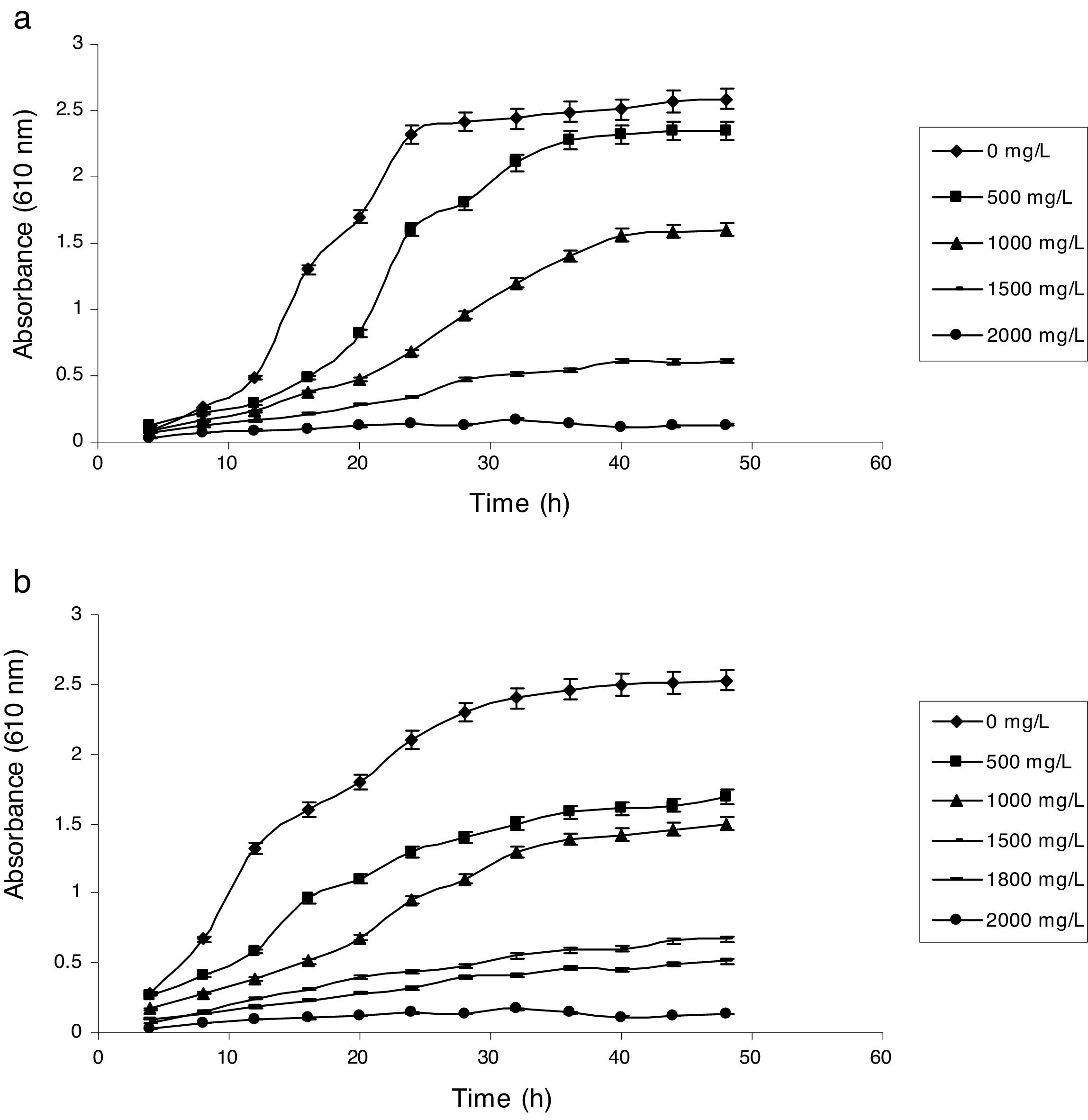
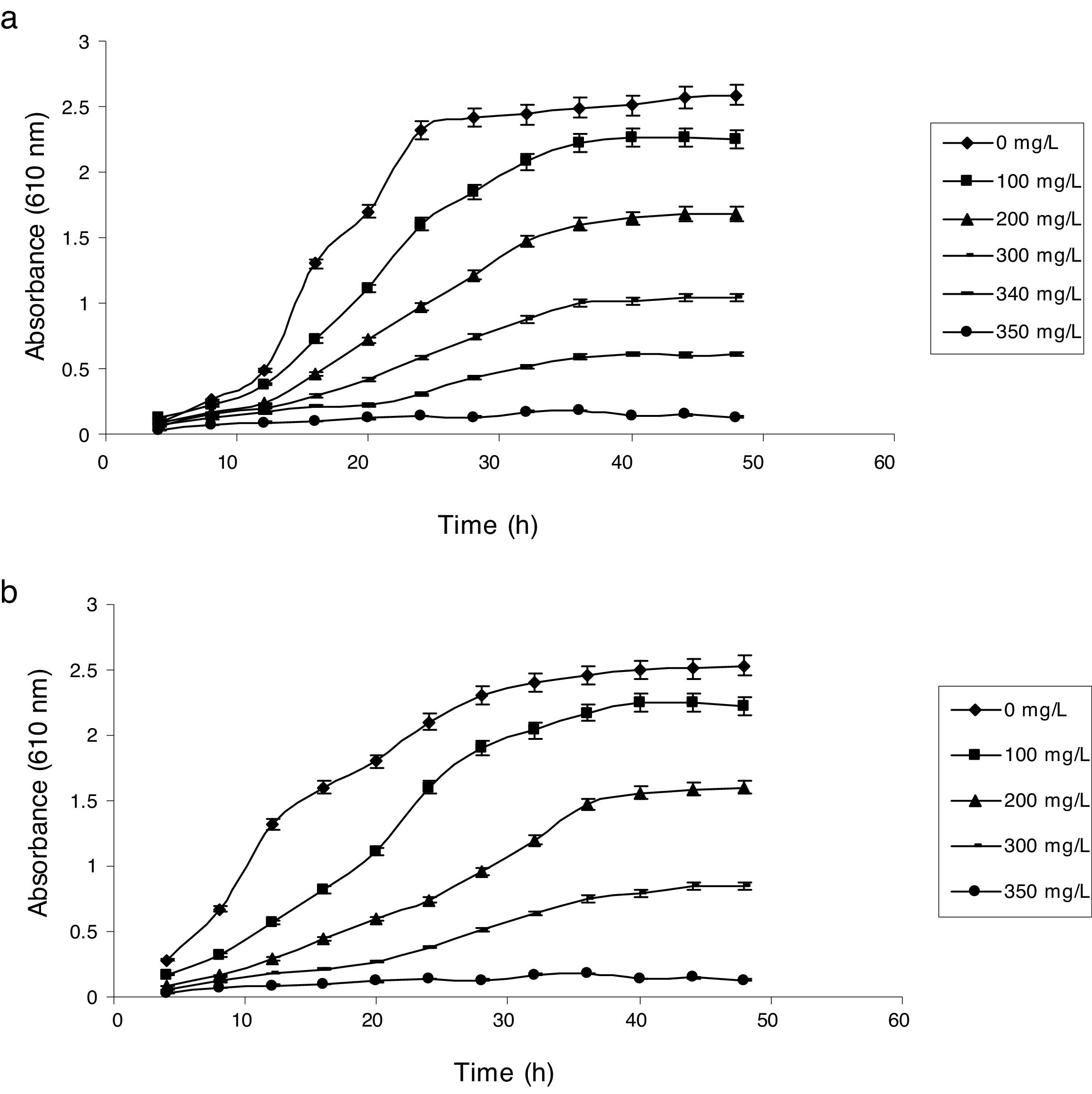
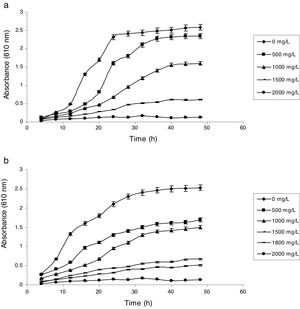
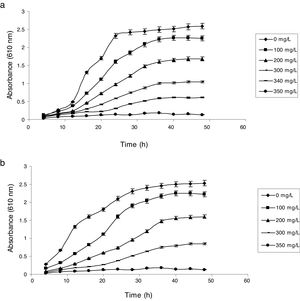
Influence of phenol and cyanide on growth of isolatesP. putida NAUN-16 tolerated phenol up to 1500mgL−1 and cyanide up to 340mgL−1 while P. stutzeri NAUN-1B tolerated phenol up to 1800mgL−1 and cyanide up to 300mgL−1 concentrations (Figs. 1 and 2). Although many reports on individual degradation of phenol and cyanide are available,18,33–37 but there are negligible reports on the simultaneous degradation of both phenol and cyanide at such high concentrations.
Effect of immobilized cells on effluentSaw dust had the maximum inherent moisture content and water-holding capacity of 16.41% and 354% respectively, followed by activated charcoal (7.02% and 246%) and fly ash (0.3% and 200%). All the carriers were near neutral in pH. The pH values of all the three solid carriers were in between 6.8 and 7.9 (saw dust, pH=6.84; charcoal, pH=7.76; fly ash, pH=7.1).
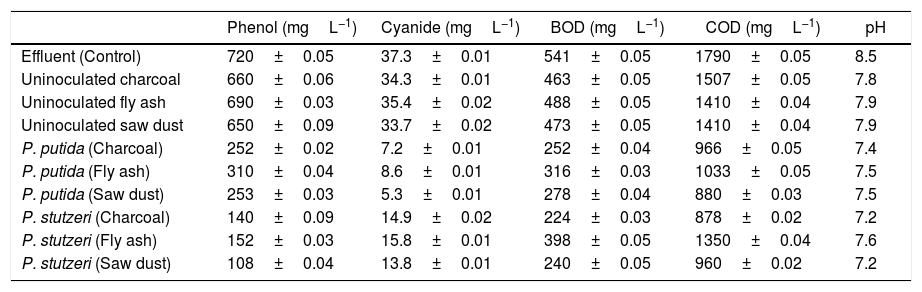
P. putida NAUN-16 immobilized on fly ash degraded 56.8% phenol and 76.9% cyanide from the effluent. It reduced BOD and COD by 41% and 42% respectively. When immobilized on charcoal it reduced phenol concentration by almost 60.8% and cyanide by 80.6% and reduced BOD and COD by 53% and 46% respectively. When immobilized on saw dust it reduced phenol by 64.8% and cyanide by 85.7%.and BOD and COD by 48% and 51% respectively. P. stutzeri NAUN-1B when immobilized on fly ash degraded 78.8% phenol and 57.6% cyanide from the effluent. It reduced BOD and COD by 26% and 23% respectively. When immobilized on charcoal it reduced phenol concentration by almost 80.5% and cyanide by 60% and reduced BOD and COD by 58% and 50% respectively. When immobilized on saw dust it reduced phenol by 84.9% and cyanide by 63% and BOD and COD by 55% and 46% respectively (Table 1).
Composition of coke-oven effluent treated with two screened bacterial isolates as compared to untreated effluent.
| Phenol (mgL−1) | Cyanide (mgL−1) | BOD (mgL−1) | COD (mgL−1) | pH | |
|---|---|---|---|---|---|
| Effluent (Control) | 720±0.05 | 37.3±0.01 | 541±0.05 | 1790±0.05 | 8.5 |
| Uninoculated charcoal | 660±0.06 | 34.3±0.01 | 463±0.05 | 1507±0.05 | 7.8 |
| Uninoculated fly ash | 690±0.03 | 35.4±0.02 | 488±0.05 | 1410±0.04 | 7.9 |
| Uninoculated saw dust | 650±0.09 | 33.7±0.02 | 473±0.05 | 1410±0.04 | 7.9 |
| P. putida (Charcoal) | 252±0.02 | 7.2±0.01 | 252±0.04 | 966±0.05 | 7.4 |
| P. putida (Fly ash) | 310±0.04 | 8.6±0.01 | 316±0.03 | 1033±0.05 | 7.5 |
| P. putida (Saw dust) | 253±0.03 | 5.3±0.01 | 278±0.04 | 880±0.03 | 7.5 |
| P. stutzeri (Charcoal) | 140±0.09 | 14.9±0.02 | 224±0.03 | 878±0.02 | 7.2 |
| P. stutzeri (Fly ash) | 152±0.03 | 15.8±0.01 | 398±0.05 | 1350±0.04 | 7.6 |
| P. stutzeri (Saw dust) | 108±0.04 | 13.8±0.01 | 240±0.05 | 960±0.02 | 7.2 |
Note: Each value is an average of five replicates ±SD.
The microorganisms were resistant to washout and toxicity because they were held on a stationary carrier surface. It has been shown that high cell concentration results in a high degradation rate of toxic compounds.38–40 It is probably due to accelerated reaction rates caused by high local cell density in or on the immobilized matrix.41 Immobilized cells offer the possibility of degrading higher concentrations of toxic pollutants than can be achieved with free cells.42,43
There was also some reduction in phenol (8.3% in charcoal; 9.7% in saw dust; 4.1% in fly ash), cyanide (8.0% in charcoal; 9.6% in saw dust; 5.0% in fly ash), BOD (14.4% in charcoal; 19.2% in saw dust; 9.7% in fly ash) and COD (16.8% in charcoal; 21.2% in saw dust; 11.4% in fly ash) with un-inoculated carriers only (Table 1). As far as the carriers were concerned saw dust showed best results for both isolates. Kulandaivel et al.44 also reported saw dust as a very good carrier. Immobilization provides a kind of membrane stabilization, which may be responsible for the protection of cells and better degradation rates.41,45
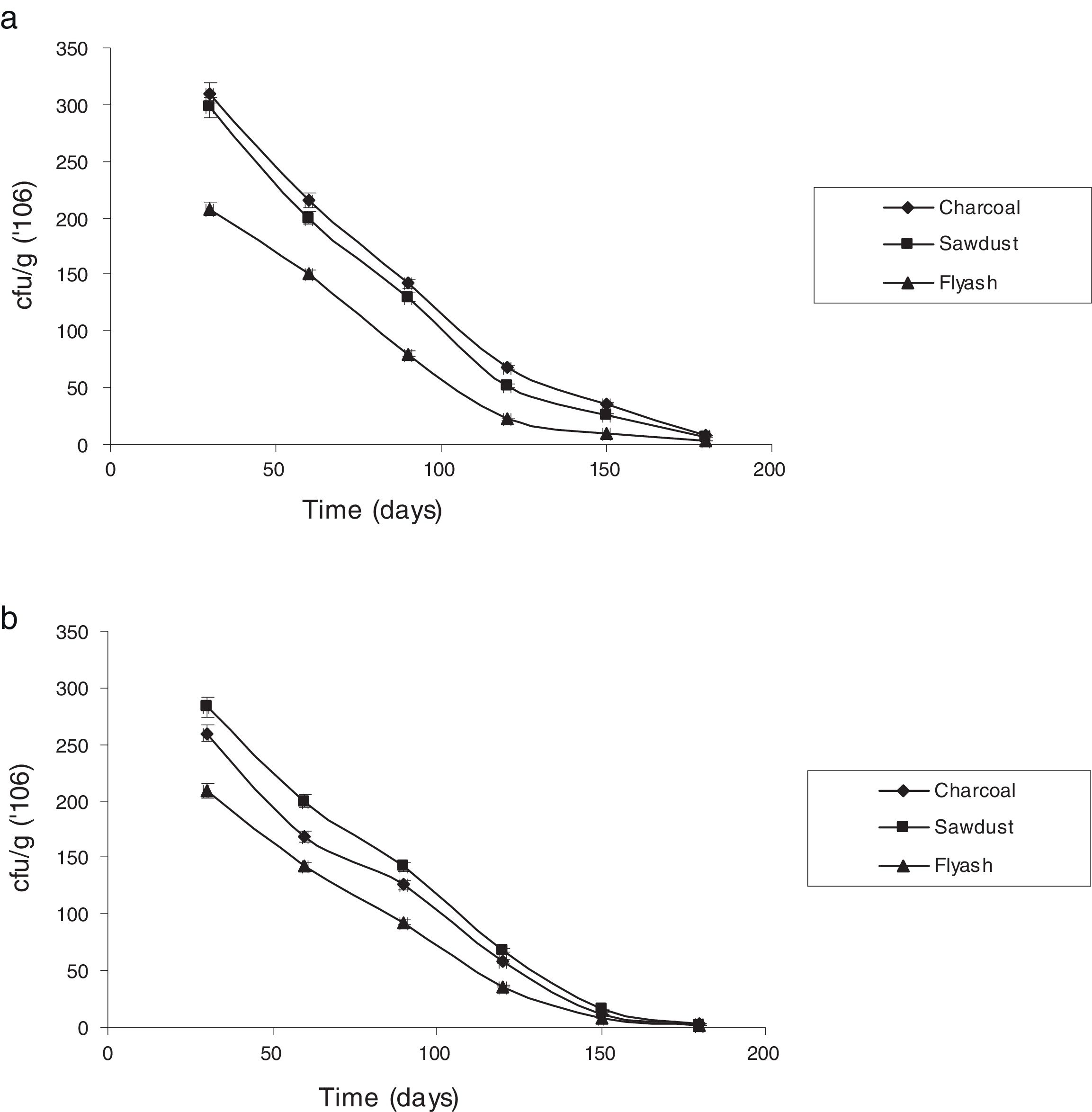
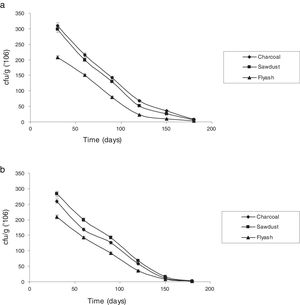
Survival rate of immobilized cells with reference to storage timeThe potential of immobilizing cells for treatment of industrial effluents is of great value.7 The stability of the immobilized cells under storage was assessed by checking viability at different times when stored at room temperature (25±5°C). Fig. 3 shows the changes in the number of viable cells of P. putida NAUN-16strep+tet+ and P. stutzeri NAUN-1Bamp+strep+. Charcoal and saw dust proved to be much better carriers. Arora et al.46 observed that carriers possessing high water-holding capacity and near neutral pH can support large populations.
Charcoal and saw dust also supported almost similar number of bacterial cells before and after the seven days of treatment process. P. putida NAUN-16strep+tet+ gave 8.49logcfug−1 before and 8.45logcfug−1 after treatment in charcoal, 8.47logcfug−1 before and 8.44logcfug−1 after treatment in saw dust and 8.31logcfug−1 before and 8.25logcfug−1 after treatment in fly ash. Likewise, P. stutzeri NAUN-1Bamp+strep+ gave 8.41logcfug−1 before and 8.36logcfug−1 after treatment in charcoal, 8.45logcfug−1 before and 8.37logcfug−1 after treatment in saw dust and 8.32logcfug−1 before and 8.25logcfug−1 after treatment in fly ash. The main advantage of using immobilized cells of microorganisms is their higher operational stability.47 Immobilization of viable cells provides a better environment for reaction and alters physiological features in metabolism such as enhanced enzyme induction. Moreover, immobilization system also provides high biomass concentrations due to the formation of biofilm.28
Phenol and cyanide are present in effluents especially those produced by heavy industries using coke-ovens. The biological degradation of coke-oven effluent becomes much more difficult due to the presence of both of these toxic compounds.48 Both the bacterial strains not only showed the capacity to grow in presence of phenol and cyanide but also degrade them concurrently, which is supposed to be a novel finding of the study.
ConclusionThe microorganisms isolated from the coke-oven effluent contaminated sites had the ability to degrade and tolerate high phenol concentration up to 1800mgL−1 and cyanide up to 340mgL−1. Two of the most tolerant isolates were identified and characterized as P. putida and P. stutzeri. The study reported the simultaneous degradation of 720mgL−1 phenol and 37mgL−1 cyanide present in the effluent. Also, the BOD and COD were found to be reduced by more than 50%. Being highly tolerant of phenol and cyanide and also having a capacity to degrade phenol and cyanide in wastewater are features that make P. putida and P. stutzeri particularly useful for treatment of coke-oven effluent containing phenol and cyanide.
The study thus reports the isolation of some very important bacteria having the ability to simultaneously degrade phenol and cyanide present in coke-oven effluent. The ability to degrade improved with simple cell immobilization process. Such strains can be used in development of microbial technology for bioremediation of waste lands where industrial effluents rich in phenol and cyanide are dumped.
Conflicts of interestThe authors declare no conflicts of interest.