Mastitis adversely affects milk production and in general cows do not regain their full production levels post recovery, leading to considerable economic losses. Moreover the percentage decrease in milk production depends on the specific pathogen that caused the infection and enterobacteria are responsible for this greater reduction. Phenotypic tests are among the currently available methods used worldwide to identify enterobacteria; however they tend to misdiagnose the species despite the multiple tests carried out. On the other hand The Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS) technique has been attracting attention for its precise identification of several microorganisms at species level. In the current study, 183 enterobacteria were detected in milk (n=47) and fecal samples (n=94) from cows, and samples from water (n=23) and milk lines (n=19). All these samples were collected from a farm in Rio de Janeiro with the specific purpose of presenting the MALDI–TOF MS technique as an efficient methodology to identify Enterobacteriaceae from bovine environments. The MALDI–TOF MS technique results matched the biochemical test results in 92.9% (170/183) of the enterobacteria species and the gyrB sequencing confirmed 100% of the proteomic technique results. The amino acid decarboxylation test made the most misidentifications and Enterobacter spp. was the most misidentified genus (76.9%, 10/13). These results aim to clarify the current biochemical errors in enterobacteria identification, considering isolates from a bovine environment, and show the importance for more careful readings of phenotypic tests which are often used in veterinary microbiology laboratories.
Brazil is the fifth largest milk producer in the world and had a production of 6128 billion liters in the first half 2015.1 Dairy livestock are present throughout Brazil contributing to the generation of employment and regional development and consequently dairy farming is important for the country's economy.2 However, environmental bovine mastitis is a major challenge in the primary sector as it causes a drop in milk production, premature discard of animals, treatment costs and has a negative effect on the quality of milk, which in turn interferes in the industrial processing of dairy products.3
Environmental mastitis is associated mainly to Gram negative bacteria, especially Enterobacteriaceae. This family of bacteria colonizes the intestine of mammals and birds and also infects the mammary glands after milking due to the contact of the udder with infected water, bedding, milking environment or cattle shed.4 The family Enterobacteriaceae has more than 50 genera and over 200 species. Escherichia, Klebsiella, Enterobacter, Serratia and Proteus are genus frequently isolated from dairy environments. Escherichia coli are naturally present in feces from warm-blooded animal species, Klebsiella, Enterobacter and Serratia marcescens inhabit soils, grains, and water. Proteus spp. usually contaminants hose water used to wash udders before milking.5,6 Thus, characterization of circulating Enterobacteriaceae in a dairy production environment is essential to understand the importance of these bacteria as agents of bovine mastitis, as well as their routes of contamination.
The quality of microbial identification can impact the veterinary clinical management becoming an essential task to control mastitis. The identification of the bacteria in routine clinical microbiology laboratories is still based on phenotypic tests, however, some strains within a species may have small biochemical differences that can result in false results in vitro.7,8 Phenotypic properties are unstable at times and their expression is dependent upon changes in the environmental conditions, e.g., growth substrate, temperature, and pH levels.9 In addition, commercially available identification systems sometimes are unable to identify some organisms.10
The Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry has been used for bacteria detection and identification using an easy to follow protocol.11 The method is based on the accurate determination of their protein mass that is compared to available profiles stored in a software database identifying the specie in a few minutes.8 MALDI–TOF MS provides fast and accurate results on species identification thus improving the overall microbiological diagnosis.12
Molecular techniques, such as 16S rRNA sequencing, are the gold standard for bacterial identification but the current bacteria characterization by these techniques is unusual and too expensive for veterinary laboratories. However, they can be used as a tool to help understand the discrepancies between the commonly used protocols. For example, the gyrB gene – encoding gyrase subunit B or topoisomerase II has been considered more discriminating than 16S rRNA, since it allows the differentiation and identification of closely related species within the Enterobacteriaceae family.
This study compared the MALDI–TOF MS technique to traditional biochemical-base methods to identify Enterobacteriaceae from bovine milk and feces, as well as in samples from water and milk lines of a dairy production system to determine the possible errors in biochemical tests. Also, the incorrect identifications were confirmed through gyrB gene sequencing.
Materials and methodsEthics statementThis study was carried out in cooperation with a dairy farmer in the municipality of Barra do Piraí, Rio de Janeiro, Brazil. This report is not intended to be a field study; instead it describes the application and accuracy of MALDI–TOF MS technique and conventional biochemical tests used in laboratories for the identification of coliform bacteria associated to bovine mastitis. Therefore, details and influence of the seasons on samples are not included. This study was conducted according to ethical standards and approved by the Ethics Committee and Biosafety of the institution under protocol number: CEUA-3664040915. The samples used in the current study were obtained from samples submitted for routine veterinary diagnosis, which was unrelated to this research. The report is focused on the microbiological analysis following the isolation of bacteria from the milk and fecal samples, and did not directly involve any of the animals.
Material collectionThe present study was performed in the town of Barra do Piraí, Rio de Janeiro, Brazil (Latitude: 22° 28′14″ South, Longitude: 43° 49′36″ West) in 2014 and 2015. A pool of milk samples from 94 cows tested positive by the California Mastitis Test (CMT) were collected, over three consecutive weeks. A total of 94 rectal feces samples from these same lactating cows were also collected. Representing milk line samples (n=48): 10 samples from workers’ hands, 10 nasal samples from workers, 20 milking machine samples and 8 nasal samples of pets (dogs and cats present in the milking parlor). Finally, one sample from each farm water supply was collected–water from well, weir, faucet, drinking fountain and brook, making a total of 19 water samples.
Phenotypic identificationThe bovine milk and milk line samples were first inoculated on blood agar (blood agar base enriched with 5% sheep blood), while the fecal and water samples were inoculated on EMB (Eosin Methylen Blue) agar and incubated at 35°C (±2°C) for 24h. Then, the isolates were submitted to routine microbiological diagnostics, including inoculation in selective medium for analysis of cultural properties. The Gram negative bacteria identification was followed by a protocol comprising: glucose and lactose fermentation with gas production, H2S (hydrogen sulfide), indole, motility, acetoin and mixed acid production, lysine and ornithine decarboxylation, arginine dehydrogenase, urease production, citrate and malonate.13 All tests were carried out in triplicate. The strains Escherichia coli ATCC25922 and Klebsiella pneumoniae ATCC700603 were used as quality control.
Proteomic identification (MALDI–TOF MS)Furthermore, all isolates were evaluated by MALDI–TOF MS. Initially, the samples were inoculated in BHI agar at 37°C for 24h. Each culture was transferred to a microplate (96 MSP, Bruker® – Billerica, USA). The bacterial sediment was covered with a lysis solution (70% formic acid; Sigma–Aldrich®). Then a 1-μL aliquot of matrix solution (alpha-ciano-4-hidroxi-cinamic acid diluted in 50% acetonitrile and 2.5% trifluoroacetic acid, Sigma–Aldrich®) was added. The spectra of each sample were generated in a mass spectrometer (MALDI–TOF LT Microflex, Bruker®) equipped with a 337nm nitrogen laser in a linear path, controlled by the FlexControl 3.3 (Bruker®) program. The spectra were collected in a mass range between 2000 and 20,000m/s, and then were analyzed by the MALDI Biotyper 2.0 (Bruker®) program, using the standard configuration for bacteria identification, which compared the spectrum of the samples with the references in the database. The results vary on a 0–3 scale, where the highest value means a more precise match and reliable identification. In this study, we accepted values for matching greater than or equal to 2.
gyrB sequencingThe bacterial DNA was extracted after thermal lysis and the 700bp gyrB fragment amplification was obtained by Polymerase Chain Reaction technique (PCR). The PCR was performed using the primers UP1 and 181r.14,15 The concentrations used for the reactions were: buffer 10× (10mM Tris–HCl, pH 9.0; 50mM KCl and 0.1% Triton X-100), 2.0mM of MgCl2, 0.5μM of each primer, 0.2mM of dNTP (Fermentas) and 0.5U of Taq polymerase (Fermentas) in a total reaction volume of 20μL with 20ng of the extracted DNA. The fragments were evaluated by electrophoresis in agarose gel (1.5%) and reveled with the SYBR green dye (Invitrogen), making visualization in ultraviolet light possible as well as being able to record the amplicons with the image capture system L-PIX EX (Loccus Biotecnologia). The PCR products were purified using an Exo-Sap (USB Corporation, Cleveland, Ohio), as recommended by the manufacturer and submitted to the sequencer ABI 3130xl of Applied Biosystems (Biotechnology Laboratory of the Genomic Sciences Post-Graduation Program of the Catholic University of Brasilia). The sequences were edited using the Bioedit program16 and Mega version 4.017 and compared with other sequences in the NCBU database (GenBank: http://www.ncbi.nlm.nih.gov/) using the BLAST algorithm.18
StatisticsThe biochemical tests and MALDI–TOF MS association were evaluated by Chi-Square test (χ2) and Fisher's test with 95% confidence interval, considering wrong identification in genera or at a specie level.
Results and discussionA total of 183 enterobacteria were detected from 443 samples: 51.3% (94/183) were from bovine feces, 25.6% (47/183) from bovine milk, 12.6% (23/183) from water and10.3% (19/183) from the milk line. All isolates were submitted to biochemical tests and proteomics and both procedures identified isolates at a specie level.
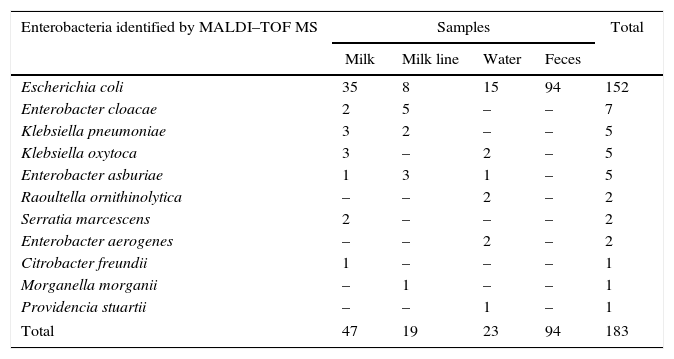
The MALDI–TOF MS identified E. coli as the prevalent specie in all samples evaluated (83%, 152/183) (Table 1). Although E. coli was isolated mainly from the feces samples it was also detected in other samples. This specie is one of the most thoroughly studied free-living organisms. It is also a remarkably diverse species because some E. coli strains live as harmless commensals in animal intestines.19 Its presence is a definite indication of fecal contamination and some strains can cause a wide variety of intestinal and extra-intestinal diseases, such as diarrhea, urinary tract infections, septicemia and mastitis.20
Enterobacteria identified by MALDI–TOF MS isolated from different samples in a bovine environment.
| Enterobacteria identified by MALDI–TOF MS | Samples | Total | |||
|---|---|---|---|---|---|
| Milk | Milk line | Water | Feces | ||
| Escherichia coli | 35 | 8 | 15 | 94 | 152 |
| Enterobacter cloacae | 2 | 5 | – | – | 7 |
| Klebsiella pneumoniae | 3 | 2 | – | – | 5 |
| Klebsiella oxytoca | 3 | – | 2 | – | 5 |
| Enterobacter asburiae | 1 | 3 | 1 | – | 5 |
| Raoultella ornithinolytica | – | – | 2 | – | 2 |
| Serratia marcescens | 2 | – | – | – | 2 |
| Enterobacter aerogenes | – | – | 2 | – | 2 |
| Citrobacter freundii | 1 | – | – | – | 1 |
| Morganella morganii | – | 1 | – | – | 1 |
| Providencia stuartii | – | – | 1 | – | 1 |
| Total | 47 | 19 | 23 | 94 | 183 |
The milk sample showed the highest variably of enterobacteria with seven of the eleven bacteria isolated. This is probably due to the methodology of the milk collection used in this study – for three consecutive weeks, pointing to possible changes in milk microbiome. However, because of the low number of samples more investigations of each cow evaluated would be necessary to confirm this hypothesis. In spite of that, the presence of a large variability of enterobacteria is worrying since these bacteria frequently are associated to clinical bovine mastitis.21
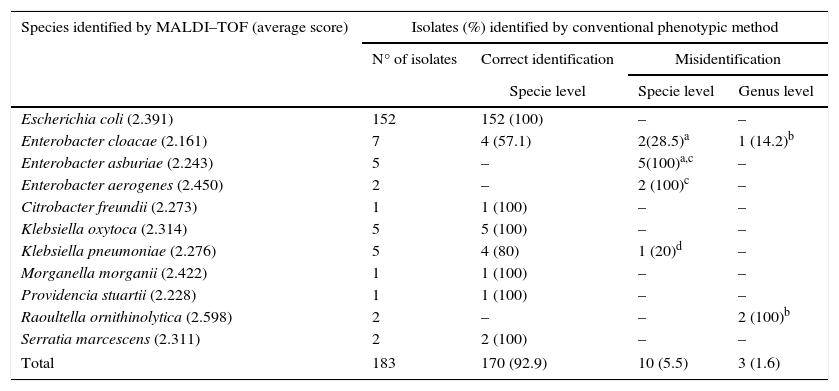
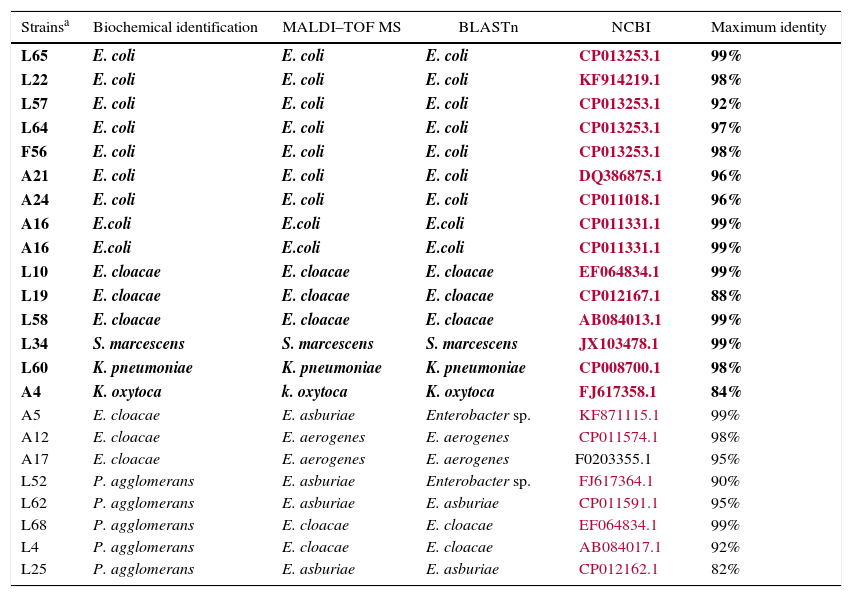
The MALDI–TOF MS technique matched with 92.9% (n=170) of the biochemical identifications results (Table 2). Other authors have also found a high correlation between these two techniques. Eigner et al. (2009)22 described 95.2% of consistency in 1116 bacterial strains previously identified in clinical routine and Bizzini et al. (2010)11 obtained correct identification of 95.1% of 1278 strains at species level and 3% at genus level. One of the major advantages of using this technology for bacterial identification is the time-to-result, which is reduced from 1 to 6 days to less than an hour. In addition, MALDI–TOF MS allowed accurate bacterial identification of a large variety of bacteria. Genotypic identification using coa, nuc and ADNr 23S genes compared to MALDI–TOF had 100% sensitivity and specificity, according to Motta et al. (2014)23 in a Staphylococcus spp. study. Dubois et al. (2010)24 and Carpaij et al. (2011)25 demonstrated that proteomic techniques are an accurate method for determining coagulase-negative Staphylococcus (CNS) species compared to molecular identification of clinical isolates. After proteomic identification, 13 (7.10%, 13/183) isolates misidentified were submitted to gyrB sequencing. There were no amplification results for five isolates as it was not possible to extract their DNA (1 K. pneumoniae, 1 E. cloacae, 1 E. asburie and 2 Raoultella ornithinolytica). Therefore, only eight enterobacteria with non-concordant identification were analyzed with this technique. Additionally, 15 isolates with concordant identification were sequenced to be the control (Table 3). The gyrB sequencing results demonstrated six species had been identified correctly by MALDI–TOF MS: two E. aerogenes (A12 and A17), two E. asburiae (L62 and L25) and two E. cloacae (L62 and L4). One isolate (L52) identified as P. agglomerans by biochemical tests and as E. asburie by proteomic test was characterized as genus Enterobacter. These results show that gyrB sequencing and the MALDI–TOF MS technique produce similar results. Another isolate identified as E. clocae and as E. asburie by biochemical and proteomic techniques, respectively, was also characterized only at genus level by sequencing. Thus, in the present study the proteomic technique was used as the “gold standard” for evaluating the sensitivity and specificity of phenotypic tests used in enterobacteria identification.
The phenotypic methods in comparison to MALDI–TOF MS in identification of enterobacteria isolated from a bovine environment.
| Species identified by MALDI–TOF (average score) | Isolates (%) identified by conventional phenotypic method | |||
|---|---|---|---|---|
| N° of isolates | Correct identification | Misidentification | ||
| Specie level | Specie level | Genus level | ||
| Escherichia coli (2.391) | 152 | 152 (100) | – | – |
| Enterobacter cloacae (2.161) | 7 | 4 (57.1) | 2(28.5)a | 1 (14.2)b |
| Enterobacter asburiae (2.243) | 5 | – | 5(100)a,c | – |
| Enterobacter aerogenes (2.450) | 2 | – | 2 (100)c | – |
| Citrobacter freundii (2.273) | 1 | 1 (100) | – | – |
| Klebsiella oxytoca (2.314) | 5 | 5 (100) | – | – |
| Klebsiella pneumoniae (2.276) | 5 | 4 (80) | 1 (20)d | – |
| Morganella morganii (2.422) | 1 | 1 (100) | – | – |
| Providencia stuartii (2.228) | 1 | 1 (100) | – | – |
| Raoultella ornithinolytica (2.598) | 2 | – | – | 2 (100)b |
| Serratia marcescens (2.311) | 2 | 2 (100) | – | – |
| Total | 183 | 170 (92.9) | 10 (5.5) | 3 (1.6) |
The gyrB sequencing results in enterobacteria identified by biochemical tests and MALDI–TOF MS.a
| Strainsa | Biochemical identification | MALDI–TOF MS | BLASTn | NCBI | Maximum identity |
|---|---|---|---|---|---|
| L65 | E. coli | E. coli | E. coli | CP013253.1 | 99% |
| L22 | E. coli | E. coli | E. coli | KF914219.1 | 98% |
| L57 | E. coli | E. coli | E. coli | CP013253.1 | 92% |
| L64 | E. coli | E. coli | E. coli | CP013253.1 | 97% |
| F56 | E. coli | E. coli | E. coli | CP013253.1 | 98% |
| A21 | E. coli | E. coli | E. coli | DQ386875.1 | 96% |
| A24 | E. coli | E. coli | E. coli | CP011018.1 | 96% |
| A16 | E.coli | E.coli | E.coli | CP011331.1 | 99% |
| A16 | E.coli | E.coli | E.coli | CP011331.1 | 99% |
| L10 | E. cloacae | E. cloacae | E. cloacae | EF064834.1 | 99% |
| L19 | E. cloacae | E. cloacae | E. cloacae | CP012167.1 | 88% |
| L58 | E. cloacae | E. cloacae | E. cloacae | AB084013.1 | 99% |
| L34 | S. marcescens | S. marcescens | S. marcescens | JX103478.1 | 99% |
| L60 | K. pneumoniae | K. pneumoniae | K. pneumoniae | CP008700.1 | 98% |
| A4 | K. oxytoca | k. oxytoca | K. oxytoca | FJ617358.1 | 84% |
| A5 | E. cloacae | E. asburiae | Enterobacter sp. | KF871115.1 | 99% |
| A12 | E. cloacae | E. aerogenes | E. aerogenes | CP011574.1 | 98% |
| A17 | E. cloacae | E. aerogenes | E. aerogenes | F0203355.1 | 95% |
| L52 | P. agglomerans | E. asburiae | Enterobacter sp. | FJ617364.1 | 90% |
| L62 | P. agglomerans | E. asburiae | E. asburiae | CP011591.1 | 95% |
| L68 | P. agglomerans | E. cloacae | E. cloacae | EF064834.1 | 99% |
| L4 | P. agglomerans | E. cloacae | E. cloacae | AB084017.1 | 92% |
| L25 | P. agglomerans | E. asburiae | E. asburiae | CP012162.1 | 82% |
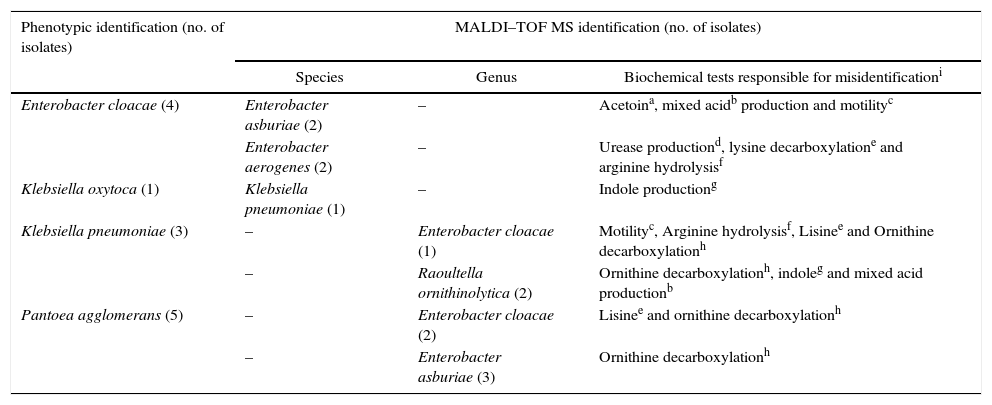
The species correctly identified by biochemical tests were E. coli (n=152), Klebsiella oxytoca (n=5), Klebsiella pneumoniae (n=4), Enterobacter cloacae (n=4), Serratia marcescens (n=2), Citrobacter freundii (n=1), Morganella morganii (n=1), Providencia stuartii (n=1). The biochemical tests that misread species identification, in this work, are presented in Table 4.
Biochemical-based tests that differ from the identification by MALDI–TOF MS.
| Phenotypic identification (no. of isolates) | MALDI–TOF MS identification (no. of isolates) | ||
|---|---|---|---|
| Species | Genus | Biochemical tests responsible for misidentificationi | |
| Enterobacter cloacae (4) | Enterobacter asburiae (2) | – | Acetoina, mixed acidb production and motilityc |
| Enterobacter aerogenes (2) | – | Urease productiond, lysine decarboxylatione and arginine hydrolysisf | |
| Klebsiella oxytoca (1) | Klebsiella pneumoniae (1) | – | Indole productiong |
| Klebsiella pneumoniae (3) | – | Enterobacter cloacae (1) | Motilityc, Arginine hydrolysisf, Lisinee and Ornithine decarboxylationh |
| – | Raoultella ornithinolytica (2) | Ornithine decarboxylationh, indoleg and mixed acid productionb | |
| Pantoea agglomerans (5) | – | Enterobacter cloacae (2) | Lisinee and ornithine decarboxylationh |
| – | Enterobacter asburiae (3) | Ornithine decarboxylationh | |
The phenotypic test parameters (sensitivity and specificity) showed high values due to their high level performance confirmed by MALDI–TOF MS. After the proteomic identification the thirteen misidentified isolates were used to investigate the mistakes occurred in the phenotypic identification. As described above fourteen tests were used in the biochemical-based identification and eight of them were detected as being responsible for wrong results; these tests were: bacterial motility in semi-solid medium, acetoin production (Voges Proskauer test), mixed acids production (Methyl Red Test), urease and indole production, arginine hydrolysis, lysine and ornithine decarboxylation.
Among the discordant results, Enterobacter spp. was the most common genus misidentified (77%, 10/13). Enterobacter asburiae (n=5) were misidentified as Pantoea agglomerans (n=3) and Enterobacter cloacae (n=2). Some E. asburie were considered false positive by the VP test but acetoin production by this specie can be expressed in only 79% of isolates which probably leads to misidentification. Pantoea agglomerans was belonging to Enterobacter genus, classified as Enterobacter agglomerans.26 It is difficult to differentiate Pantoea spp. from the other members of this family, such as the Enterobacter, Klebsiella and Serratia species. Recently, six species were assigned to the Enterobacter cloacae complex, including Enterobacter asburiae, Enterobacter cloacae, Enterobacter hormaechei, Enterobacter kobei, Enterobacter ludwigii, and Enterobacter nimipressuralis. In surveillance studies, Enterobacter species are not often classified beyond the genus level, probably because identification is difficult. Only the Enterobacter spp. isolates that belong to the E. cloacae complex are of clinical significance and are increasingly isolated as human pathogens. Thus, more precise identification of the E. cloacae complex isolates may permit differentiation between pathogens, commensal and transitional species. Up to now phenotypic identification of species and subspecies within the E. cloacae complex have been largely unreliable and irreproducible.27
Two isolates of Raoultella ornithinolytica were misidentified as Klebsiella pneumoniae. The indole, VM and ornithine decarboxylation tests are responsible to differentiate R. ornithinolytica and K. pneumoniae and probably were mistakenly interpreted. Historically, these species are closely related. Drancourt et al. (2001)28 using phylogenetic analysis of rpoB genus confirmed the heterogeneity of Klebsiella spp. and proposed the formation of three groups: Group I comprise three subspecies, K. pneumoniae subsp. pneumoniae, K. pneumoniae subsp. ozaenae and K. pneumoniae subsp. rhinoscleromatis; Group II comprise K. terrigena, K. ornithinolytica, K. planticola and K. trevisanii; and Group III is represented by K. oxytoca. Based on this evidence, these authors proposed a division of Klebsiella genus: Klebsiella spp. and Raoultella spp. All Group II species were transferred to the new genus Raoultella spp. In the present study Raoultella ornithinolytica was isolated from water samples. This species is often recovered from water, soil, plants and occasionally mammal mucosa, and so may be associated to mastitis.23
One isolate of Klebsiella pneumoniae was misidentified at the genus-level. By proteomic technique it was identified as Enterobacter cloacae. Although belonging to a different genus it is included in the same tribe – Klebsielleae, along with Hafnia, Serratia and Pantoea genera. In this study, Klebsiella pneumoniae presented false negative results for arginine, ornithine and motility tests as well as false positive to lysine. The amino acids decarboxylation tests are extremely important, especially for the separation of certain members of the Klebsiella-Enterobacter-Serratia-Hafnia group.13
Another K. pneumoniae was misidentified as K. oxytoca. The indole production could differentiate them but some strains do not produce classic reactions leading to designation of several additional species and consequently inducing the error of identification.29 And the misidentification of E. aerogenes as E. cloacae was probably due to the urease interpretation reading for E. cloacae, which is variable in most cases.13
Considering all the tests used in the present study, the lysine decarboxylation presented the lowest sensitivity percentage (81%). Interpretation of the tests for decarboxylation of amino acids must be accurate especially because some isolates present a slow reaction resulting in a wrong identification. Additionally, the bacterial density, the incubation time and the pH properly adjusted are required standards to minimize the risk of false results.25 In general, microorganisms first ferment glucose to lower the pH so that the optimal hydrogen ion concentration for decarboxylase activity is reached. Decarboxylation results in the formation of amines and consequently increases the pH. Positive results are usually obtained after 18–24h of incubation but with some strains the tests must be held for as long as 4 days. Oxygen is generally excluded by overlaying the broth with paraffin or mineral oil or by the addition of 0.3% agar.30,31 A medium with agar cannot be held longer than 24h and thus a few false negative results should be expected. A large number of microbiologists have adopted the use of the lysine Falkow broth instead of the Möller agar, because the Falkow test eliminates the anaerobic or acid environment. However, this test cannot be used to detect the lysine decarboxylase activity of certain members of the Klebsiella-Enterobacter-Serratia-Hafnia group considering the acetyl methyl carbinol production which interferes at the end of alkaline pH change leading to a false negative interpretation.13
The bacterial motility test induced misidentification of two isolates (E. cloacae and E. asburiae). Some authors suggest the use of semi-solid medium with tetrazolium salts to evaluate the motility. This salt was converted into red and insoluble complexes by the reducing properties of most enterobacteria thus improving the visualization.32 In the same way, the indole and mixed acids production tests also depend on individual viewing making this test subjective in most cases. The subjectivity of phenotypic tests has increased the search for more definitive techniques in bacterial identification such as molecular tools.9 But, equally important with regard to these molecular methods is the scientific accuracy of the biochemical identification of each species since clinical and veterinary laboratories continue to rely upon the biochemical properties and not upon 16S rDNA gene sequencing to identify bacterial strains.
ConclusionIn the present study, the MALDI–TOF MS technique results matched in 92.9% with the phenotypic test results and the gyrB sequencing confirmed the proteomic results in 100%. E. coli was prevalent in all samples collected from the bovine environment and the milk bovine sample presented the highest microbiota variation. The E. coli specie was easily identified by MALDI–TOF MS while Enterobacter spp. was the most misidentified genus. The phenotypic test parameters showed high values (sensitivity>81% and specificity>89%) due to their high level performance confirmed by the proteomic technique; however eight of them were associated with misidentification at species or genus levels. The amino acid decarboxylation test was the cause of the majority of the misidentifications. This shows the importance for more careful readings of phenotypic tests which are often used in veterinary clinical microbiology laboratories.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by the National Council for Scientific and Technological Development (CNPq, Rio de Janeiro, Brazil – process 308528/2011-5) and Foundation for Research Support in the State of Rio de Janeiro (FAPERJ).