Sophora tomentosa is a pantropical legume species with potential for recovery of areas degraded by salinization, and for stabilization of sand dunes. However, few studies on this species have been carried out, and none regarding its symbiotic relationship with beneficial soil microorganisms. Therefore, this study aimed to evaluate the diversity of nitrogen-fixing bacteria isolated from nodules of Sophora tomentosa, and to analyze the occurrence of colonization of arbuscular mycorrhizal fungi on the roots of this legume in seafront soil. Thus, seeds, root nodules, and soil from the rhizosphere of Sophora tomentosa were collected. From the soil samples, trap cultures with this species were established to extract spores and to evaluate arbuscular mycorhizal fungi colonization in legume roots, as well as to capture rhizobia. Rhizobia strains were isolated from nodules collected in the field or from the trap cultures. Representative isolates of the groups obtained in the similarity dendrogram, based on phenotypic characteristics, had their 16S rRNA genes sequenced. The legume species showed nodules with indeterminate growth, and reddish color, distributed throughout the root. Fifty-one strains of these nodules were isolated, of which 21 were classified in the genus Bacillus, Brevibacillus, Paenibacillus, Rhizobium and especially Sinorhizobium. Strains closely related to Sinorhizobium adhaerens were the predominant bacteria in nodules. The other genera found, with the exception of Rhizobium, are probably endophytic bacteria in the nodules. Arbuscular mycorrhizal fungi was observed colonizing the roots, but arbuscular mycorhizal fungi spores were not found in the trap cultures. Therefore Sophora tomentosa is associated with both arbuscular mycorhizal fungi and nodulating nitrogen-fixing bacteria.
Due to their ability to establish symbiosis with nodulating nitrogen-fixing bacteria (NNFB) and arbuscular mycorrhizal fungi (AMF), some species of the Fabaceae family are able to grow in unfavorable environments. Generally, individuals of this family are used as pioneers in soil conservation and to recover degraded areas, since they enrich the soil with organic matter with high nitrogen, and consequently allow the establishment of other plants in the ecological succession process. According to Nobrega et al.,1 soil salinization is one of the most serious forms of soil degradation. Areas of saline soils are found in the shores of oceans, lakes, and in arid and semi-arid agricultural areas, due to intensive use of irrigation with water of high salts content, and more recently, in areas of industrial landfill.2 Plant species adapted to high salt concentrations are able to develop morphophysiological mechanisms that make it possible for them to grow and develop in these conditions.3,4 In this context, studying species able to settle in salty conditions and understanding their mechanisms and adaptations have been a challenge for agricultural scientists and for those interested in recovering degraded areas.2
The legume Sophora tomentosa L., locally known in Rio de Janeiro as “feijão da praia”, “comandaiba” or “rosario”, is likely one of those species adapted to saline environments because they are usually found in seafront soils subjected to intermittent salination from seawater. In Brazil, it occurs naturally in sandbanks from the northeast to the south of the country, and its natural occurrence has been reported in the Pacific Islands,5 Oceania,6 East Asia3 and Mexico.7 However, Peña et al.8 claim that this species has been found in the coast of all tropical regions of the world, characterizing it as a species of pantropical distribution. It is a shrub plant, about 3 m high, with fast growth and high aggressiveness, forming large clumps and offering good coverage to the ground. Its roots are abundant, with a volume about three times greater than the shoot.9 The fructification is practically continuous, and it blooms throughout the year, with sporadic interruptions.10 Its capacity of nodulation was observed when it was inoculated with Sinorhizobium fredii11 and Sinorhizobium adhaerens12 under controlled conditions, but only the second formed nitrogen-fixing nodules.
Despite its potential for recovery of degraded and salinized areas, and also for containment of dunes,2 few studies on this species are found in the literature. Even less works regarding their symbiotic relationships with beneficial microorganisms, such as rhizobia and mycorrhizal fungi, especially at natural conditions are found. Therefore, this study aimed to evaluate the diversity of nodulating nitrogen-fixing bacteria from nodules of S. tomentosa roots, and to analyze the occurrence of colonization of arbuscular mycorrhizal fungi in the roots of this legume, with native inocula from a Spodosol.
Material and methodsCharacterization of the sampling areaThe Municipal Ecological Park Prainha (MEPP), located in the western part of Rio de Janeiro (RJ), between the coordinates 23°01′52″–23°02′30″ S and 43°30′00″–43°30′38″ W, has Aw climate (tropical with rains in summer), according to the Köppen classification. The average annual rainfall is between 1200 and 2000mm, and the average temperature varies from 18°C in winter to 32°C in summer. According to Veloso et al.,13 the native vegetation is classified as Sub-mountain Dense Rain Forest (on the slopes) and Pioneer Formations (sandbank and wetlands). S. tomentosa is found on the sandy beach, together with heliophytic and halophytes species, such as “capim-de-praia” (Sporobolus virginicus – Graminae), “feijão de praia” (Carnavália rosea – Fabaceae), locally scarce “guriri” (Allagoptera arenaria – Palmae), and “abaneiro” (Clusia fluminensis – Guttiferae). Wetlands soils were classified as Spodosols, according to the Brazilian Soil Classification System.
Collection of nodules and seeds of S. tomentosa and soil sampling in the fieldSoil samples were collected for chemical and physical analysis, according to the methods compiled by Embrapa,14 which was carried out in the laboratories of the Federal University of Lavras.
Nodules (Supplementary Material – Figure S2) and seeds of S. tomentosa were collected when plants were at flowering and seed production stages. Roots of three plants were sampled, and all the nodules were collected. These nodules were packed in Nasco® sterile plastic bags and then dried in glass tubes with cotton and silica gel. Seeds were stored in paper bags and preserved in cold chamber at 4°C. To produce soil inoculum, 5 composite samples of rhizosphere soil of S. tomentosa were collected, which were stored in Nasco® sterile plastic bags at 4°C, until spores extraction. Part of these soil samples were used to capture rhizobia.
Installation of trap culture to capture rhizobia and to evaluate the colonization of AMF and spores extraction under controlled conditionsS. tomentosa seeds, from the Municipal Ecological Park Prainha were scarified in concentrated sulfuric acid, and the surface was disinfected by immersion in 1% sodium hypochlorite (v/v) for 3min, followed by successive washes with sterile distilled water. Seeds were germinated in Petri dishes with filter paper and moistened cotton, and put in an oven at 28°C. After the development of the radicles, three seedlings were transferred to 500mL plastic pots with substrate, composed of 150mL soil inoculum and 350mL sterile sand and vermiculite in a 2:1 ratio, respectively. In order to capture native AMF, Brachiaria decumbens were grown together with S. tomentosa in the same pot. For this, forage seeds were scarified in concentrated sulfuric acid for 1min, and planted directly into plastic pots, with density of four seeds per plot. Pots were kept in a greenhouse of the Department of Soil Science of the Federal University of Lavras for six months, and plants were regularly irrigated with a modified Hoagland solution,15 as described by Guimarães et al.16 After this period, three plants of S. tomentosa were collected for analysis of root nodules and presence of AMF structures. For analysis of root colonization by AMF, it was used the coloring method of root fragments.17 After dyeing the roots, they were transferred to Petri dishes and were visualized in a stereoscopic microscope with 10 to 40 times magnification. Manual cuts were carried out in some nodules in order to capture images, as well as fungal structures within S. tomentosa roots, using the optical microscope Olympus model BX40.
AMF spores extraction was carried out by the technique of wet sieving18 and centrifugation in sucrose.19
Isolation and characterization of bacterial strainsNNFB were isolated from nodules collected in the field and from trap plants. Nodules were first immersed in ethyl alcohol (95%) for 30s in order to break surface tension. After that, nodules were immersed in hydrogen peroxide (H2O2) (30%) for 3min to desinfect their surface, and then, they were washed six times with sterile distilled water to remove H2O2 excess. After that, they were crushed in Petri dishes containing culture medium 7920 using a sterilized tweezer. Petri dishes were incubated at 28°C until the appearance of isolated colonies. All identified colony morphotypes were picked and incubated at 28°C repeatedly to obtain pure cultures. Subsequently, phenotypic characterization of strains was carried out. The analyzed characteristics were: time to the appearance of isolated colonies (1–3 days – fast growth; 4 days – intermediate growth; 6 days or more days – slow growth), colony diameter (mm), changes in culture medium pH (acidification, neutralization, or alkalinization), shape (circular or irregular), colony elevation, border appearance, color (yellow, orange, white, beige, or salmon), and the consistency of the exopolysaccharides produced (gummy, aqueous, or dry), and absorption of the culture medium indicator.
From the data of phenotypic characterization, a hierarchical group analysis was carried out using the coefficient of Gower21 as dissimilarity metric and Ward algorithm22 for grouping in R.23 To define homogeneous groups on dendrogram, the criterion of Mojena was used.24
16S rRNA gene sequencingFor the 16S rRNA gene sequencing, representative isolates from each group formed in the dendrogram of phenotypic characteristics were selected. Genomic DNA extraction, amplification and sequencing of 16S rRNA gene were carried out following the methodology described by Guimarães et al.16
The quality of the sequences was verified by BioNumerics 7.6 (Aplied Maths, Sint-Martens-Latem, Belgium). The resulting sequences, with exception of RIOP243II and RIOP243, were assembled into contigs and compared with those available in the GenBank (National Center for Biotechnology Information, NCBI), using Basic Local Alignment Search Tool (BLAST). The sequence alignment was performed with 1115 base pairs using ClustalW. A phylogenetic tree was generated using MEGA version 6.025 with default parameters, Kimura 2-parameter distance model,26 and Neighbor-Joing algorithm.27 Statistic support for the tree was evaluated by bootstrap confidence analysis with 1000 samplings. Sequences of the isolates of S. tomentosa and at least two closely-related type/reference strains available in GenBank were included, with the purpose of evaluating whether these strains would form separate clusters with known species.
The sequences presented in this work were deposited in GenBank under accession numbers KY820810 to KY820830.
Authentication experimentsTwo experiments were performed aiming to authenticate the isolated bacterial strains in S. tomentosa growing in Leonard vessels filled with sand and vermiculite 1:1 and Hoagland's nutrient solution, but both of them were unsuccessful due to the poor growth of S. tomentosa in axenic conditions even after 6 months of cultivation.
ResultsSoil analysisChemical and physical analysis of soil samples from the rhizosphere of S. tomentosa is found in Table 1.
Chemical and physical analysis of Spodosol from Sophora tomentosa rhizosphere collected in the Municipal Ecological Park Prainha, RJ.
| pH in H2O | P | K | Na | Ca | Mg | Al | H+Al | SB | cec | CEC | E.C. | PST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mgdm−3 | cmolcdm−3 | mScm−1 | % | |||||||||
| 7.9 | 31.44 | 36.51 | 56.20 | 0.54 | 0.23 | 0.00 | 0.53 | 0.86 | 0.86 | 1.39 | 0.56 | 17 |
| m | V | O.M. | Prem | Zn | Fe | Mn | Cu | B | S | Sand | Silt | Clay |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | mgL−1 | mgdm−3 | % | |||||||||
| 0.00 | 62.13 | 0.09 | 56.81 | 6.99 | 64.75 | 10.19 | 0.99 | 0.03 | 4.35 | 100 | 0 | 0 |
SB: Sum of bases (Ca2++Mg2++K+); cec: cation exchange capacity in natural pH; CEC: cation exchange capacity at pH=7.0; E.C.: electrical conductivity; PST: percent of sodium saturation; m: aluminum saturation; V: base saturation at pH=7.0; O.M.: organic matter.
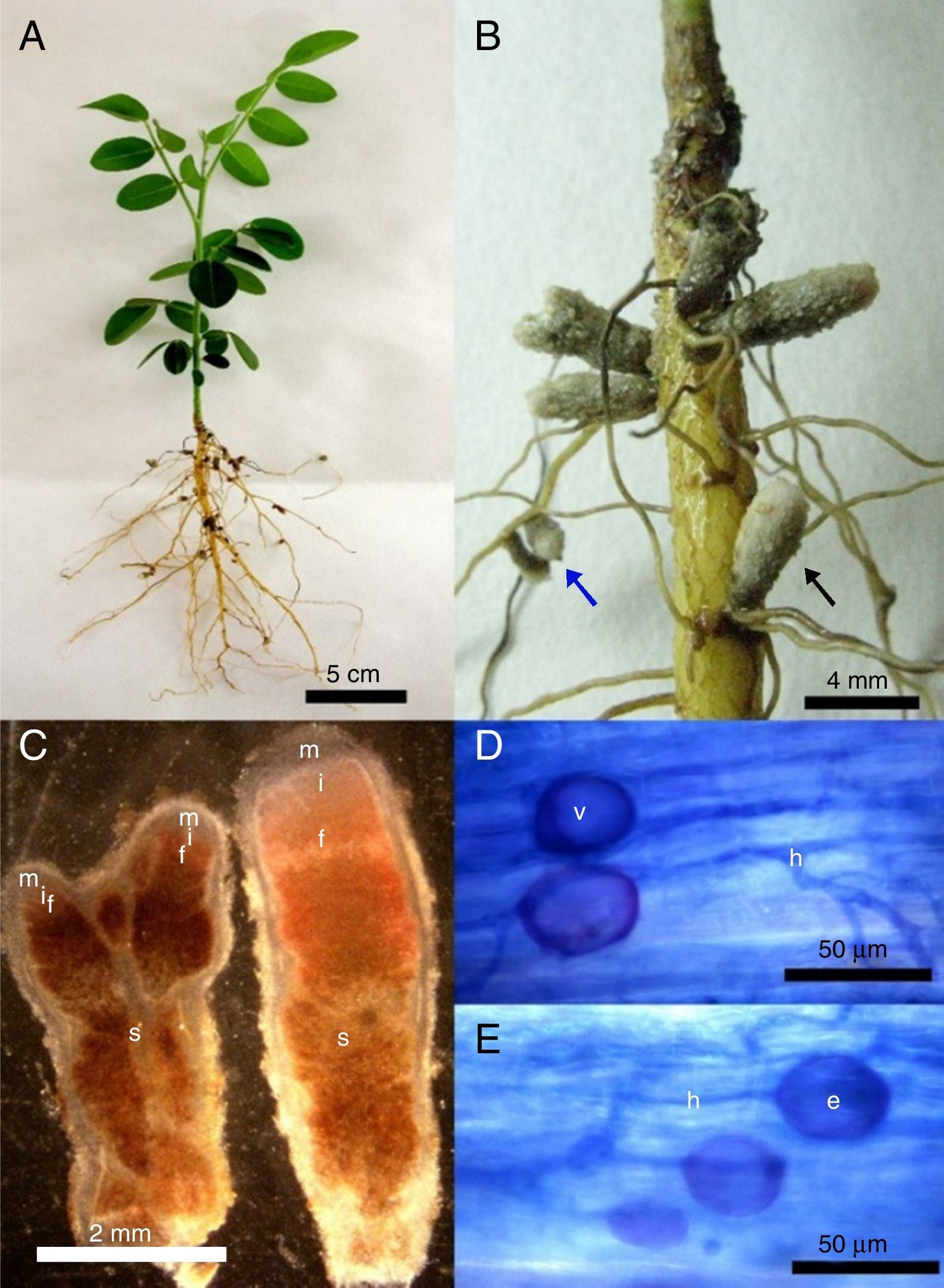
After six months of cultivation, using S. tomentosa as trap plant for NNFB capture, it was observed that plants grew very slowly, reaching only about 10cm, and did not develop the root system well (Fig. 1A). However, plants were nodulated in this period, and nodules with indeterminate growth and redish color inside (Fig. 1B) occurred throughout the root. The observed nodules had length ranging from 1 to 6mm, and contained one or two apical meristems, suberized surface and darkening when senescent (Fig. 1B and C).
(A) Plant appearance after being cultivated for 6 months; (B) nodules with indeterminate growth with suberized surface occurring in lateral roots base (black arrow) or along them (blue arrow). (C) Longitudinal section of nodules with one (right) or two (left) apical meristems (m), presenting senescent regions (s), fixation (f), infection and differentiation zones (i). (D–E) Roots colonized by arbuscular mycorrhizal fungal with intra radicular spores (e), vesicles (v) and fungal hyphae (h).
From the analysis of root colonization by AMF, it was observed the presence of typical structures, such as coenocytic hyphae, vesicles and intra radicular spores in the roots (Fig. 1D and E). However, only two spores were found in all extractions carried out from the field samples and trap cultures.
Capture, isolation and phenotypic characterization of strainsA total of 20 nodules were selected from the trap cultures (12) and the field (8). As a result, 36 bacterial strains were isolated from the nodules collected in the field, and 15 from the trap culture (Fig. 2). According to the phenotypic characteristics of the strains, five groups were separated according to the homogeneity criterion of Mojena.24 Of these groups, 21 representatives strains were chosen randomly and had their DNAs sequenced, being 2 strains from the first group, 2 from the second, 13 from the third, 2 from the fourth and 2 from the fifth group. These 21 strains showed rapid growth; 16 strains acidified the culture medium, 3 did not change the pH, and 2 alkalinized the culture medium (Table 2).
Cultural characteristics and identification of the bacterial strains from Sophora tomentosa nodules.
| Strain | Cultural group | CCa | BPb | Most similar sequence found in the genbank | ||
|---|---|---|---|---|---|---|
| Accesion number | % of similarity | Species | ||||
| RIOP231 | 3 | RAC | 1253C | AJ420773 | 100 | Sinorhizobium/Ensifer adhaerens |
| RIOP243II | 3 | RAC | 774F | KU877657 | 100 | Sinorhizobium/Ensifer adhaerens |
| RIOP243III | 3 | RAC | 1248C | AJ420773 | 100 | Sinorhizobium/Ensifer adhaerens |
| RIOP243V | 3 | RN | 1002F | AJ420773 | 100 | Sinorhizobium/Ensifer adhaerens |
| RIOP245I1 | 3 | RAC | 1246C | AJ420773 | 100 | Sinorhizobium/Ensifer adhaerens |
| RIOP245I22 | 3 | RAC | 1287C | AJ420773 | 100 | Sinorhizobium/Ensifer adhaerens |
| RIOP245II1 | 3 | RAC | 1124C | AJ420773 | 100 | Sinorhizobium/Ensifer adhaerens |
| RIOP245II22 | 3 | RN | 1203C | AJ420773 | 100 | Sinorhizobium/Ensifer adhaerens |
| RIOP245III | 1 | RAC | 1226C | AJ420773 | 100 | Sinorhizobium/Ensifer adhaerens |
| SIIIP3N1 | 1 | IAC | 1203C | NR115768 | 99 | Sinorhizobium mexicanus |
| NR116265 | 99 | Sinorhizobium chiapanecum | ||||
| RIOP321 | 3 | RAC | 1267C | KT380607 | 100 | Rhizobium pusense |
| RIOP322 | 3 | RAC | 1276C | KT380607 | 100 | Rhizobium pusense |
| RIOP323 | 3 | RAC | 1244C | KT380607 | 100 | Rhizobium pusense |
| RIOP324 | 3 | RAC | 1256C | KT380607 | 100 | Rhizobium pusense |
| RIOP325 | 3 | RAC | 1254C | KT380607 | 100 | Rhizobium pusense |
| RIOP243IV | 4 | RAL | 1220C | KT580622 | 100 | Brevibacillus brevis |
| KT719605 | 100 | Brevibacillus formosus | ||||
| SIIP1N1I | 4 | RAL | 1297C | NR112926 | 99 | Brevibacillus nitrificans |
| NR040980 | 99 | Brevibacillus choshinensis | ||||
| RIOP344 | 2 | RAC | 1401C | GU985441 | 99 | Paenibacillus kribbensis |
| RIOP251I1 | 2 | RAC | 1310C | KY407752 | 100 | Bacillus aryabhattai |
| RIOP251II | 5 | RAC | 1405C | KY407752 | 100 | Bacillus aryabhattai |
| RIOP346 | 5 | RN | 1306C | KU156696 | 100 | Bacillus cereus |
| KY003095 | 100 | Bacillus thuringiensis | ||||
Cultural characteristics growth and change of pH: RAC, rapid growth and acidic pH; RAL, rapid growth and alkaline pH; RN, rapid growth and neutral pH; IAC, intermediate growth and acid pH.
Number of base pairs in the 16S RNA sequence used for identification: F, foward; C, Contig. Strains with prefix “RIO” were isolated from nodules collected in the field and the prefix “SI” from the nodules collected in trap plants cultivated in axenic condition and inoculated with soil from the rhizosphere.
From the 16S rRNA gene sequence and comparisons with sequences available in the GenBank, 11 known bacterial species were most closely related to the isolates from nodules of S. tomentosa (Table 2). These results are presented in the phylogenetic tree constructed with 16S rRNA gene sequences (Fig. 3). Among the strains, seven formed a homogeneous cluster with S. adhaerens. Besides, they were well separated from the next phylogenetically closest Sinorhizobium species (S. morelense), suggesting their classification as Sinorhizobium adherens (RIOP231, RIOP243II, RIOP245I1, RIOP245I22, RIOP245II1, RIOP245II22, and RIOP245III). The strain SIIP3N1 grouped with three known species within Sinorhizobium sp; as such, our data only support its classification at the genus level. Four strains formed a separate cluster with Rhizobium pusense (RIOP321, RIOP322, RIOP323, RIOP324, RIOP325) and did not group with any other known species within the Rhizobium genus, supporting their classification as R. pusense. RIOP346 was identified as Bacillus sp., whereas RIOP251I1 and RIOP251II were closely and exclusively associated with Bacillus aryabhattai (RIOP251I1 and RIOP251II). RIOP243IV and SIIP1N1I were classified as Brevibacillus sp.; and RIOP344, as Paenibacillus kribensis.
The group of gram-negative bacteria (Rhizobium sp. and Sinorhizobium spp.) make up about 68% (13 of 19) of the strains and gram-positive (Bacillu spp., Brevibacillus spp. and Paenibacillus sp.) the other 32% (Fig. 3).
DiscussionSoil analysisAccording to the Manual of Liming and Fertilization of the state of Rio de Janeiro28 for N2-fixing legume trees and shrubs, the levels of P and K were classified as appropriate and low, respectively. The highest concentration of P can be related to the influence of seawater in the soil, enriching it with this nutrient in readily available form to plants,29 or the presence of marine molluscs shells composing sand texture, which are rich in calcium phosphate. Similar results were also find by Lourenço Junior and Cuzzuol,30 studying the influence of the soil on chemical composition of Passiflora mucronata and Canavalia rosea in a similar ecosystems.
Although the soil analysis does not indicate saline conditions in the soil at the time of sampling, these ecosystems are subject to intermittent influence of seawater and occasional salinization. However, because of the sandy texture, the ions are easily leached with percolating water after heavy rainfalls, common in the area.
Morphology of S. tomentosa nodules and AMF colonizationTransverse and longitudinal sections of the nodules showed reddish color inside, indicating the presence of Leghemoglobin (Fig. 1C) and nitrogen-fixing activity. Nodules that do not internally have this substance express whitish shades and demonstrate the inability of microsymbionts to fix nitrogen.31
Despite the observation of arbuscular mycorrhizal fungi colonization in the roots of S. tomentosa, few spores were found in soils in all extractions carried out from the field samples and trap cultures. In soil samples from the field, results may be related to the sandy texture and to the frequent soil water saturation. Moreover, the high amount of water in this environment can be a limiting factor for sporulation.32
Diversity of bacterial strains isolated from S. tomentosaIn the group of gram-negative bacteria, two distinct groups were found. The first composed by Sinorhizobium spp; the second, with Rhizobium spp. The isolates grouped with S. adhaerens type strain (Fig. 3) presented 100% similarity with this species (Table 2) and did not group with any other species of this genus, suggesting that these strains belong to S. adhaerens. A similar result was also found by Hung et al.12 in Taiwan, who identified this species as the unique symbiont of S. tomentosa. This interesting relationship between S. tomentosa and S. adhaerens found in distant locations of the globe can be an indicative of a specific symbiotic association, and their co-occurrence in distinct continents suggest that the bacteria is dispersed with S. tomentosa seeds and fruits, through the seawater.9 The bacteria could infect the seeds right after they fall on the soil, and then been carried to long distances by the ocean, setting a symbiotic association in different places around the world.
In the literature, the nomenclature for S. adhaerens is not well defined. According to Willems et al.,33 it should be classified within the Sinorhizobium genus, since it is widely known as a plant symbiont, but also due to the etymology of the word, and to the great number of nitrogen-fixing representatives within the genus Sinorhizobium, when compared to the Ensifer genus. In a more recent research, Ormeño-Orrillo et al.34 demonstrated that Ensifer adhaerens species showed enough phylogenetic difference to allocate itself in a separate group from the other species of this genus, as evidenced by Willems et al.33
In the second group of gram negative bacteria, Rhizobium spp, it is clear that five strains (RIOP321, RIOP322, RIOP323, RIOP324 and RIOP325) formed a subgroup 100% similar (Table 2) to R. pusense (Fig. 3). There is little information on R. pusense, and it was described by Panday et al.35 for isolates from the rhizosphere of Cicer arietinum L.
Gram-positive bacteria, such as species of the genera Bacillus, Paenibacillus (syn. Bacillus), and Brevibacillus were also found among the sequences obtained. Although bacteria of these genera are not known to nodulate legumes, they have been commonly isolated from nodules of various legume species and considered to be endophytic.36–40
The genus Bacillus is a phenotypically large and diverse group of Gram-positive or Gram-variable, spore-forming bacteria. In this group, representatives adapted to different environmental conditions are found. Due to advances in molecular biology, this genus has been subjected to considerable reclassification, with high phylogenetic heterogeneity.41–43 In our work, the strain RIOP346 presented 100% similarity to two species of Bacillus spp (Bacillus thuringiensis and Bacillus cereus) (Fig. 3). Thus, it was grouped into Bacillus spp. On the other hand, RIOP251I1 and RIOP251II, formed one group with Bacillus aryabhattai (100% similar) and no other group, supporting its classification as B. aryabhattai. The RIOP344 strain is 99% similar to Paenibacillus kribbensis. This species is a potassium and phosphorus solubilizing bacteria, as well as nitrogen-fixing, which is widely used in agricultural production in China.44 The Paenibacillus genus was created by Ash et al.45 to accommodate a group of the genus Bacillus, and its name derived from the latin term “paene”, meaning almost, or almost Bacillus.46
Brevibacillus genus is derived from the reclassification of the strains initially allocated as Bacillus brevis, and currently includes 20 species.43 The strain SIIP1N1I was grouped together with Brevibacillus spp., but showed 99% similarity to Brevibacillus nitrificans and Brevibacillus choshinensis. The strain RIOP243IV was 100% similar to Brevibacillus brevis and Brevibacillus formous.47 Therefore, in both situations it was not possible to reliably classify them at species level; so they were identified as Brevibacillus sp.
Thus, our study indicates that S. tomentosa nodules present several endophytic species, and suggests that Sinorhizobium/Ensifer adhaerens establishes symbiosis with S. tomentosa in natural conditions, confirming previous reports in the literature.12
ConclusionsS. tomentosa legume establishes symbiosis with nitrogen-fixing bacteria, most likely with Sinorhizobium/Ensifer adhaerens species, and with mycorrhizal fungi under natural conditions of seafront in Brazil.
Bacterial species of the genera Bacillus, Brevibacillus, and Paenibacillus were found within S. tomentosa nodules as endophytic species.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank CNPq, CAPES and FAPEMIG funding agencies for the master's, doctoral and post-doctoral scholarships, and CNPq for the research productivity fellowship granted to FMS Moreira.