As a glacier retreats, barren areas are exposed, and these barren areas are ideal sites to study microbial succession. In this study, we characterized the soil culturable bacterial communities and biochemical parameters of early successional soils from a receding glacier in the Tianshan Mountains. The total number of culturable bacteria ranged from 2.19×105 to 1.30×106CFUg−1dw and from 9.33×105 to 2.53×106CFUg−1dw at 4°C and 25°C, respectively. The number of culturable bacteria in the soil increased at 25°C but decreased at 4°C along the chronosequence. The total organic carbon, total nitrogen content, and enzymatic activity were relatively low in the glacier foreland. The number of culturable bacteria isolated at 25°C was significantly positively correlated with the TOC and TN as well as the soil urease, protease, polyphenoloxidase, sucrase, catalase, and dehydrogenase activities. We obtained 358 isolates from the glacier foreland soils that clustered into 35 groups using amplified ribosomal DNA restriction analysis. These groups are affiliated with 20 genera that belong to six taxa, namely, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Actinobacteria, Bacteroides, and Deinococcus-Thermus, with a predominance of members of Actinobacteria and Proteobacteria in all of the samples. A redundancy analysis showed that the bacterial succession was divided into three periods, an early stage (10a), a middle stage (25–74a), and a late stage (100–130a), with the total number of culturable bacteria mainly being affected by the soil enzymatic activity, suggesting that the microbial succession correlated with the soil age along the foreland.
Over the past 100 years, the average global temperature has increased by 0.74°C,1 and one consequence of this temperature increase is that glaciers are retreating in many mountainous areas of the world. As the glaciers retreat, the newly exposed land is a new habitat for microorganisms2 derived from the air, clouds, snow, rain, and runoff waters from the glacier body in addition to autochthonous microorganisms.3 Bacterial communities may be key determinants of glacier foreland ecosystem stability and function because of their important roles in soil development, biogeochemical cycles and heterotrophic activities. Microbial communities change along the soil age gradient of a glacial foreland. Sigler et al.4 found that the number of dominant organism types and community evenness decreased with succession, but others found that the phylotype number, diversity, and evenness increased over time.2,5 However, most of those studies are focused on Polar and European mountain areas; therefore, studies on the bacterial community, soil biochemical properties and the correlation between bacteria and soil biochemical properties along chronosequences in such high Asian regions as the Tianshan Mountains are still needed.
The Tianshan No. 1 glacier is located in the Eastern Tianshan Mountains of Central Asia, mountains that are surrounded by desert.6 The climate in this area is a classical continental climate, and wind is an important climatic factor in the upper elevations of the mountains.7 The Tianshan No. 1 glacier has been studied intensively from a glaciological point of view since 1959,8–10 when the Tianshan Glaciological Station was built. Because of the availability of extensive glaciological data and detailed glacier retreat data, this area is an ideal location for the study of microbial distribution and growth related to both climatic and other environmental factors.11,12 Although the study of microorganisms in this area is very important, few studies have been performed. Bai et al.13 reported the bacterial diversity from permafrost in the Tianshan Mountains, and Yang et al.14 studied the permafrost bacterial and archaeal community structures in the same area. Sheng et al.15 first described the indigenous endophytic bacteria within subnival plants of the Tianshan Mountains. Wang et al.16 reported microbial biomass and soil enzyme activity variations along chronosequences, and Wu et al.11 used pyrosequencing to analyze the bacterial diversity along chronosequences. However, studies regarding the variation of the culturable bacterial communities and biochemical characteristics along chronosequences in the Tianshan Mountains are not available, and thus fundamental knowledge on the culturable bacterial communities and biochemical characteristics in the Tianshan No. 1 glacier forelands is lacking.
In this study, we present data regarding the soil biochemical properties and diverse bacteria cultured using samples from the Tianshan No. 1 glacier forelands. The results could lead to a better understanding of the initial colonization and succession patterns of microorganisms and soil development and the correlation between bacteria and soil biochemical properties along chronosequences in a high Asian region. Our aims were (1) to investigate the soil biochemical properties and culturable bacterial abundance variations along a chronosequence, (2) to determine the culturable bacterial community variations by using a low-nutrient medium cultured at 4°C and 25°C, and (3) to examine the correlations between the abundance of culturable bacteria and the soil biochemical properties with increasing soil age.
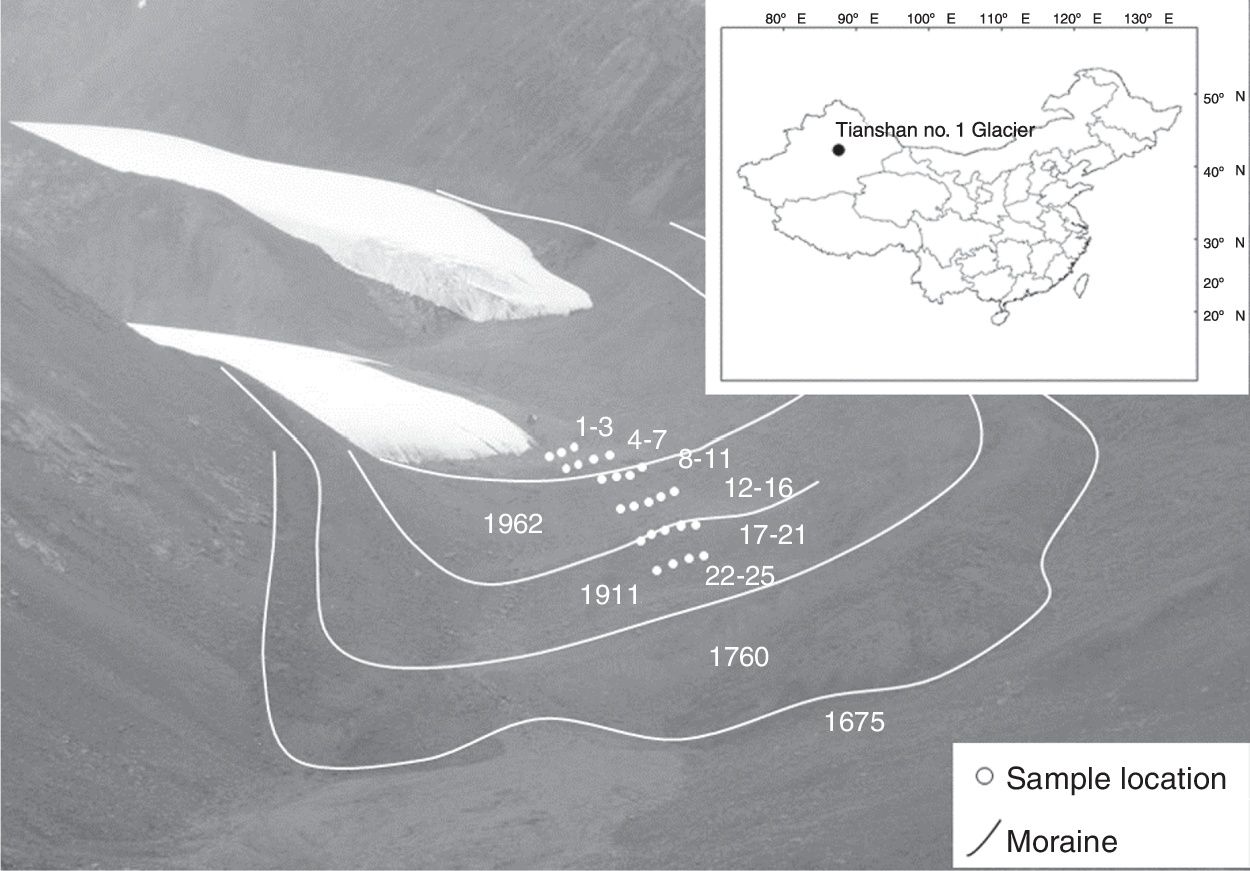
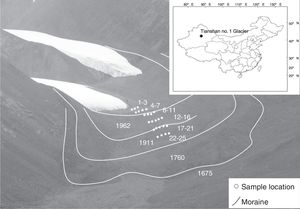
Materials and methodsStudy site and samplingThe Tianshan Mountains extend through China, Kyrgyzstan and Kazakhstan in Central Asia and have 15,953 glaciers with a total area of 15,416km2.17 The sample sites were located at Tianshan No. 1 glacier (N 43°06′, E 86°48′), 120km southwest of Urumchi, China (Fig. 1). The top elevation at this glacier is 4486m. The samples were collected at the east branch of Tianshan No. 1 glacier foreland along the chronosequence in front of the retreating glaciers. Twenty-five soil samples were collected in August 2010. These soil samples represent 6 periods: sites 1 to 3, sites 4 to 7, sites 8 to 11, sites 12 to 16, sites 17 to 21, and sites 22 to 25 represent 10a, 25a, 60a, 74a, 100a, and 130a, respectively. The succession time of every sampling site was determined using the annual glacier retreat observation data (from 1959 to 2010) from the Tianshan Glaciological Station (Chinese Academy of Sciences) and lichenometric chronology data (from 1958 to 1538).18 Each soil sample consisted of three subsample cores at a 5cm depth collected at random in an area approximately 2m×2m; the samples were mixed after the larger gravel had been removed. Pioneer plants appeared in the deglaciated soil within 10–100 years, and vegetation developed after 100 years of deglaciation. Successional species arriving within 10–100 years included Cancrinia tianschanica, Bryophyta spp., Poa tianshanica, Draba nemorosa, Saxifraga hirculus L., Melandrium apricum, Leontopodium lentopodioides, Saussurea gnaphalodes, Crepis flexuosa, Rhodiola coccinea, Oxyria digyna, and Saussurea involucrata, whereas Senecio thianschanicus, Polygonum viviparum, and Pedicularis spp. additionally appeared outside the glacier foreland. The soil samples were placed in a sterile soil box and kept on ice during transport to the laboratory and then analyzed immediately.
Biochemical analyses of the soilsThe soil water content was measured by a weight loss method after 48h at 80°C in a drying oven. The soil pH of soil: double-distilled water (1:1w/v) was measured using an acidity meter (Sartorius PT-10, Germany).19 After air-drying and grinding to allow passage through a 100 mesh sieve, the soil organic C and total N were determined using an elemental analyzer (Elementar Vario-EL, Germany).
All of the enzymatic preparations were performed at 0–4°C. The soil enzymes urease, sucrase, protease, polyphenol oxidase, catalase, and dehydrogenase were measured. The activity of urease was measured according to a modified method described by Taylor et al.,20 and the urease activity was expressed as 1mg ammonia-N/g dry weight (dw) soil per 24h. The dehydrogenase activity was determined by the reduction of 2-p-iodo-nitrophenyl-phenyltetrazolium chloride (INT) to iodo-nitrophenyl formazan (INTF) using the method of Taylor et al.,20 and the activity was expressed as 1μg INTF/g dw soil per 24h. The catalase activity was determined by measuring the O2 absorbed by KMnO4 after the addition of H2O2 to the reaction mixture.21 The protease activity was measured according to Nannipieri et al.22 and Garcıa-Gil et al.,21 and the activity was expressed as 1mg N/g dw soil per 24h. The sucrase activity was determined according to the method by Schinner,23 and the activity was expressed as 1mg glucose/g dw soil per 24h. The polyphenol oxidase activity was measured according to Saiya-Cork et al.,24 and the activity was expressed as 1mg pyrogallol/g dw soil per 24h.
Bacterial isolation and cultivationBecause a cultivation strategy using a nutrient-rich medium is rather selective, favoring fast- over slow-growing bacterial species,25 a low-nutrient medium, PYGV (DSMZ medium 621, http://www.dsmz.de), was used to isolate the bacteria. The bacterial cultivation and isolation were performed according to the method of Zhang et al.19 Soil samples (5g) were placed in a 250mL flask containing 45mL of autoclaved 0.85% NaCl solution with glass beads and shaken at 150rpm at 4°C for 30min. After this treatment, 1mL of suspended cell and soil particle subsamples were diluted from 10-fold to 10−4 with autoclaved 0.85% NaCl solution. A 100μL aliquot of the dilution was plated on PYGV medium and incubated at 25°C for one week or 4°C for three weeks. The inoculation procedure was prepared in triplicate for each dilution. A sterilized soil sample that had been autoclaved (121°C for 2h) was used as a negative control. Subsequently, the colony forming units (CFU) were calculated as the averages of the triplicate plates. Distinct colonies on the plates were restreaked onto PYGV agar plates and then immediately preserved at −70°C in liquid medium with 15% glycerol.
Amplified ribosomal DNA restriction analysis (ARDRA)ARDRA was used to group the 358 isolates. The genomic DNA was extracted and purified using an AxyPrep Bacterial Genomic Miniprep Kit (AXYGEN, USA) according to the manufacturer's instructions. Approximately 20ng of DNA was amplified with 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′), as described by Zhang et al.,19 in a total volume of 50μL. The reaction contained 2 units of Taq DNA polymerase (Fermentas), 1× Taq Buffer, 3mM MgCl2, 0.2mM dNTP, and 0.4μM each primer. After an initial step at 94°C for 3min, 30 cycles of 94°C for 1min, 58°C for 1min, and 72°C for 1.5min were performed, with a terminal 10min extension at 72°C.
A 10μL aliquot of the PCR products was double digested with restriction enzymes HaeIII and AluI (MBI Fermentas) by 1.5U of each restriction enzyme and 1μL of 10× Tango Buffer at 37°C for 16h. The enzymes were inactivated by heating the preparations at 65°C for 20min, and the products were separated using 2.5% agarose gel (wt/vol) electrophoresis in TAE buffer containing 0.5μg/mL of ethidium bromide. A 50bp ladder was used as a marker.
Sequence and phylogenetic analysisBased on the amplified ribosomal DNA restriction analysis (ARDRA) of the isolates, one representative strain of each group was selected for sequence determination of the 16S rRNA gene. Three universal primers, 27F (5′-AGAGTTTGATCCTGGCTCAG-3′), 517F (5′-CCAGCAGCCGCGGTAAT-3′), and 907F (5′-AAACTCAAATGAATTGACGGG-3′), were utilized for sequencing.19
The 16S rRNA gene sequence classifications were identified using the 16S rRNA training set 16 reference databases with the RDP Classifier method (http://rdp.cme.msu.edu/) and aligned against representative reference sequences of the most closely related members obtained from the 16S rRNA database in NCBI. CLUSTALW 1.81 software was then used to perform the multiple alignment26 using the method of Jukes and Cantor to calculate the evolutionary distances. The phylogenetic dendrograms were constructed using the neighbor-joining method,27 and the tree topologies were evaluated by performing bootstrap analysis of 1000 data sets using the MEGA4.1 package.28
Data analysisThe correlation coefficients (R) and their p values were calculated by the Pearson method using SPSS 13.0 (SPSS Inc., Chicago, IL, USA). The significance levels were within confidence limits of 0.05 or less. The data presented are the means of at least three independent experiments and are expressed as the mean±SE. Comparisons between the mean values were performed using the least significant difference (LSD test) at p<0.05. The number of each different type of colony appearing on the plates was counted when the CFU were calculated. Combining the ARDRA and sequence analysis results, the bacterial taxa abundances were determined. The analysis of culturable bacterial taxa abundance combined with the environment parameters, such as pH, C, N, and soil enzyme activity, was performed using CANOCO (version 4.5, Microcomputer Power, Ithaca, NY, USA). An initial detrended correspondence analysis (DCA) of culturable bacterial taxa abundance revealed that the data exhibited a linear (Lengths of gradient <3), rather than a unimodal, trend, which indicated that redundancy analysis (RDA) should be used.29 The total number of culturable bacteria and taxa abundance in the different aged soils were clustered by using the hierarchical cluster method (SPSS 13.0).
Nucleotide sequence accession numbersThe 16S rRNA gene sequences of the 35 representative isolated strains were deposited in GenBank. The sequences from the bacterial libraries were assigned to accession numbers: JN662509-JN662543.
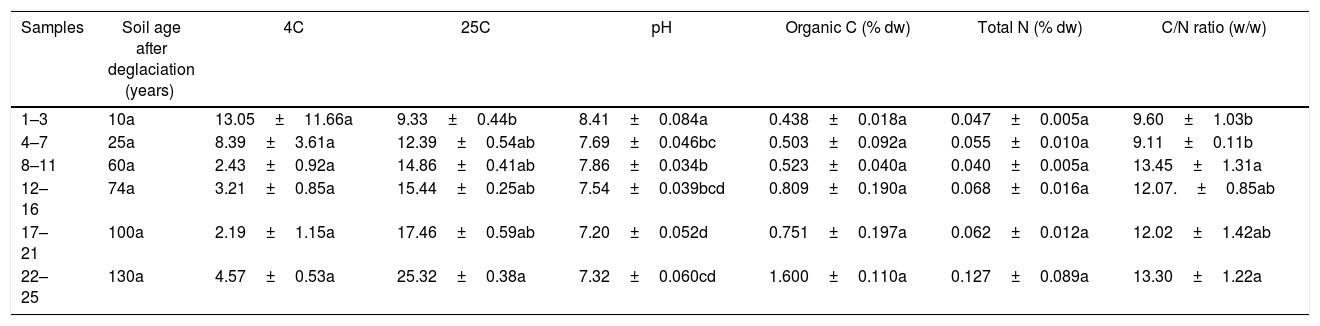
ResultsThe abundance of culturable bacteria in the soilsThe total number of the culturable bacteria ranged from 2.19×105 to 1.30×106CFUg−1dw and from 9.33×105 to 2.53×106CFUg−1dw at 4°C and 25°C, respectively. The total number of culturable bacteria isolated at 4°C decreased along the chronosequence, but not significantly (p=0.09), whereas the number of bacteria isolated at 25°C significantly increased along the chronosequence (p=0.003). The number of culturable bacteria in the 10a soil isolate at 4°C was 1.4 times that at 25°C; at later ages, the number isolated at 4°C was lower than at 25°C. At the 130a age, the number isolated at 25°C was 5.5 times that isolated at 4°C (Table 1).
The number of culturable bacteria and soil biochemical parameters of the successional sites along the chronosequences.
| Samples | Soil age after deglaciation (years) | 4C | 25C | pH | Organic C (% dw) | Total N (% dw) | C/N ratio (w/w) |
|---|---|---|---|---|---|---|---|
| 1–3 | 10a | 13.05±11.66a | 9.33±0.44b | 8.41±0.084a | 0.438±0.018a | 0.047±0.005a | 9.60±1.03b |
| 4–7 | 25a | 8.39±3.61a | 12.39±0.54ab | 7.69±0.046bc | 0.503±0.092a | 0.055±0.010a | 9.11±0.11b |
| 8–11 | 60a | 2.43±0.92a | 14.86±0.41ab | 7.86±0.034b | 0.523±0.040a | 0.040±0.005a | 13.45±1.31a |
| 12–16 | 74a | 3.21±0.85a | 15.44±0.25ab | 7.54±0.039bcd | 0.809±0.190a | 0.068±0.016a | 12.07.±0.85ab |
| 17–21 | 100a | 2.19±1.15a | 17.46±0.59ab | 7.20±0.052d | 0.751±0.197a | 0.062±0.012a | 12.02±1.42ab |
| 22–25 | 130a | 4.57±0.53a | 25.32±0.38a | 7.32±0.060cd | 1.600±0.110a | 0.127±0.089a | 13.30±1.22a |
Results are given as the means±SE. Different suffix letters indicate values that are significantly different from one another (ANOVA, p<0.05). 4C and 25C denote the number of culturable bacteria at 4°C and 25°C (×105), respectively.
The soil pH values decreased significantly (p=0.04) from 8.41 to 7.32 with the chronosequence. The soil organic C and total N content were very low at all ages of the retreating glacier, ranging from 0.438% to 1.600% and from 0.040% to 0.127%, respectively. The soil organic C significantly increased (p=0.03) along the chronosequence, and the soil total N also increased along the chronosequence, but not significantly (p=0.07). The soil C/N ratio showed an increasing trend with the soil age (p=0.06).
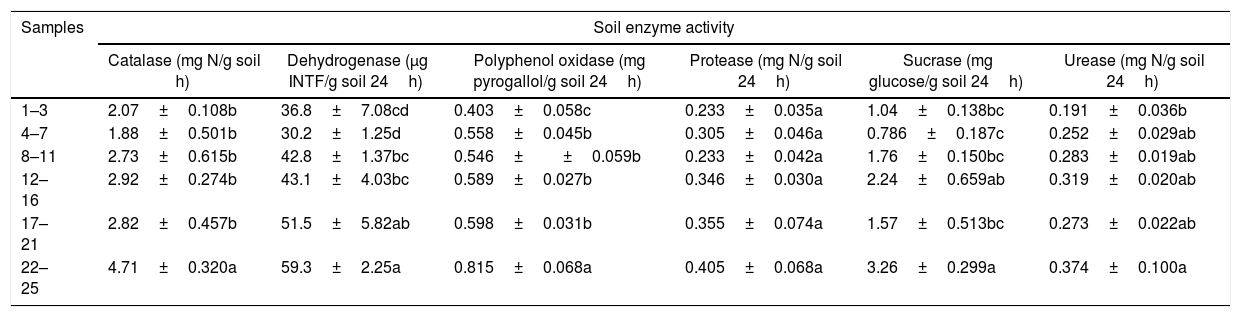
Soil enzyme activities integrate information about microbial status and soil physicochemical conditions30 and are often used as an indicator of the functioning of soil ecosystems.31 The soil enzyme activity significantly increased along the soil age gradient, including catalase, dehydrogenase, polyphenoloxidase, protease, sucrase and urease (p=0.015, 0.004, 0.017, 0.045, 0.027, and 0.023, respectively) (Tables 1 and 2).
The changes in enzyme activities in the soils of the successional sites along the chronosequences.
| Samples | Soil enzyme activity | |||||
|---|---|---|---|---|---|---|
| Catalase (mg N/g soil h) | Dehydrogenase (μg INTF/g soil 24h) | Polyphenol oxidase (mg pyrogallol/g soil 24h) | Protease (mg N/g soil 24h) | Sucrase (mg glucose/g soil 24h) | Urease (mg N/g soil 24h) | |
| 1–3 | 2.07±0.108b | 36.8±7.08cd | 0.403±0.058c | 0.233±0.035a | 1.04±0.138bc | 0.191±0.036b |
| 4–7 | 1.88±0.501b | 30.2±1.25d | 0.558±0.045b | 0.305±0.046a | 0.786±0.187c | 0.252±0.029ab |
| 8–11 | 2.73±0.615b | 42.8±1.37bc | 0.546±±0.059b | 0.233±0.042a | 1.76±0.150bc | 0.283±0.019ab |
| 12–16 | 2.92±0.274b | 43.1±4.03bc | 0.589±0.027b | 0.346±0.030a | 2.24±0.659ab | 0.319±0.020ab |
| 17–21 | 2.82±0.457b | 51.5±5.82ab | 0.598±0.031b | 0.355±0.074a | 1.57±0.513bc | 0.273±0.022ab |
| 22–25 | 4.71±0.320a | 59.3±2.25a | 0.815±0.068a | 0.405±0.068a | 3.26±0.299a | 0.374±0.100a |
Results are given as the means±SE. Different suffix letters indicate values that are significantly different from one another (ANOVA, p<0.05).
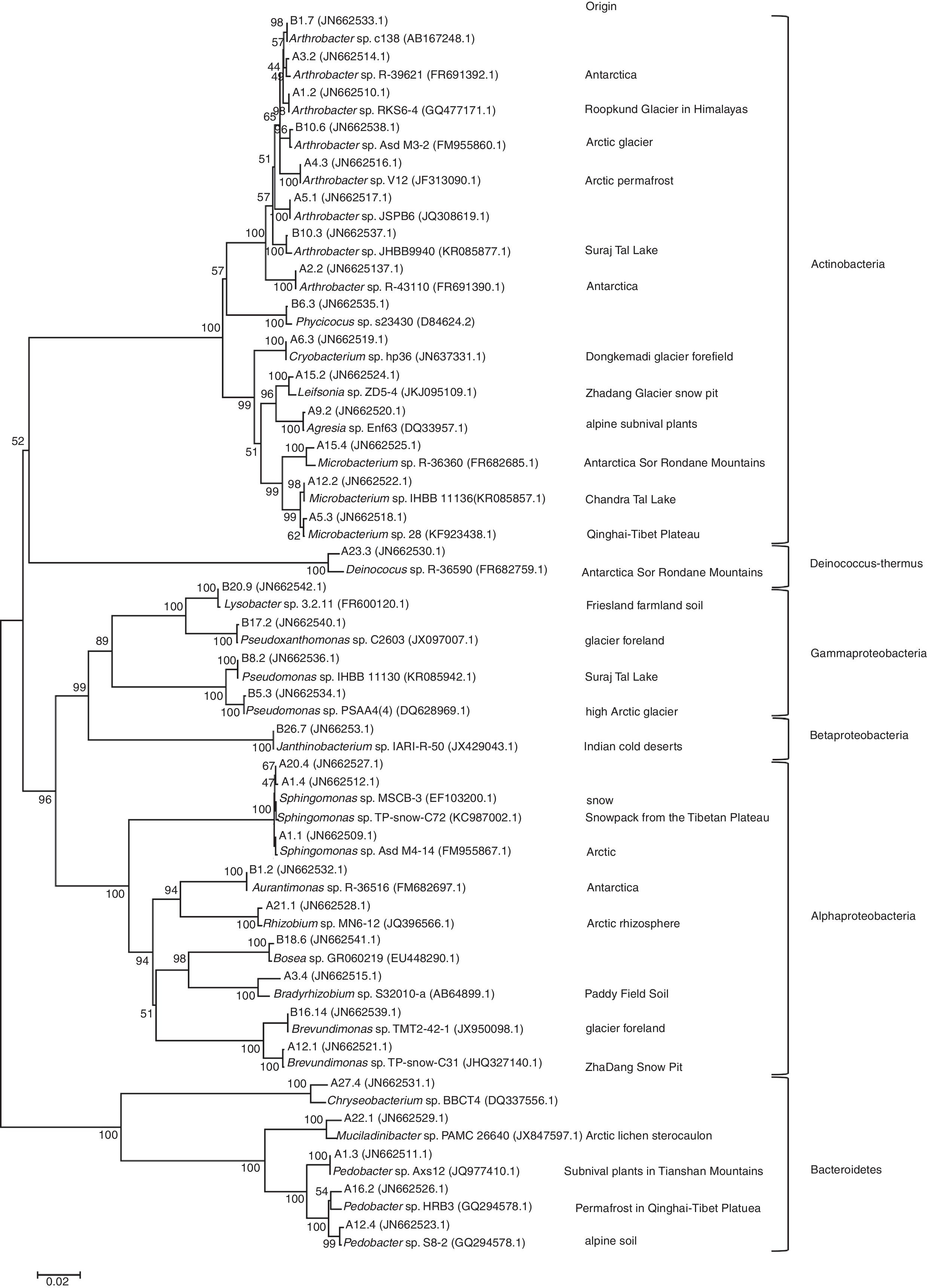
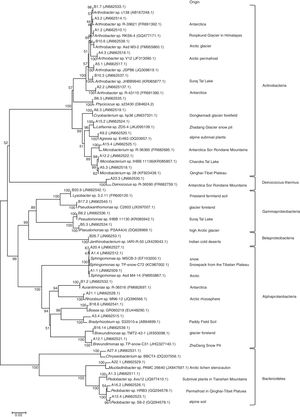
Using ARDRA, all 358 isolates cultured at 25°C and 4°C were clustered into 35 groups. After comparing the 16S rRNA sequence of a representative strain from each group, the recovered bacteria clustered into six groups: Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Actinobacteria, Bacteroides, and Deinococcus-Thermus (Fig. 2).
Phylogenetic dendrogram based on a comparison of the 16S rRNA gene sequences of the bacterial isolates from the Tianshan No. 1 glacier foreland and some of their closest phylogenetic relatives. The numbers on the tree indicate the percentages of bootstrap sampling derived from 1000 replications. The isolation source column lists the environments from which the closest phylogenetic relatives come.
The 35 studied isolates belonged to the following 20 genera: Agreia, Arthrobacter, Aurantimonas, Bosea, Bradyrhizobium, Brevundimonas, Chryseobacterium, Cryobacterium, Deinococcus, Janthinobacterium, Leifsonia, Lysobacter, Microbacterium, Mucilaginibacter, Pedobacter, Phycicoccus, Pseudomonas, Pseudoxanthomonas, Rhizobium, and Sphingomonas. A total of eight isolates belonged to Arthrobacter, and three isolates belonged each to Microbacterium, Pedobacter, and Sphingomonas. Brevundimonas and Pseudomonas each had two isolates, and the other genera were represented by one isolate each. As revealed by the phylogenetic tree construction, these bacteria were closely related to other cold-environment bacteria (Fig. 2).
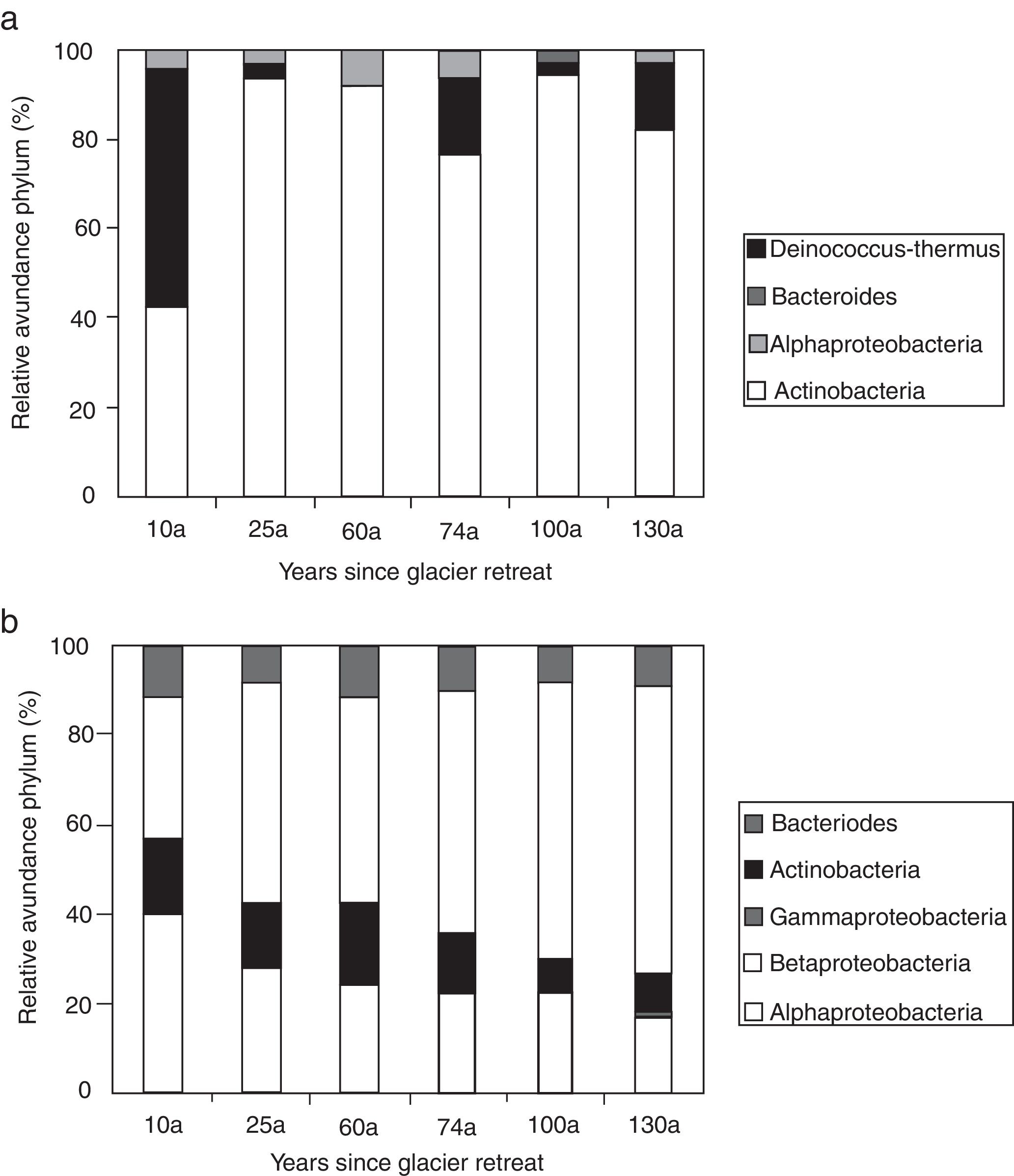
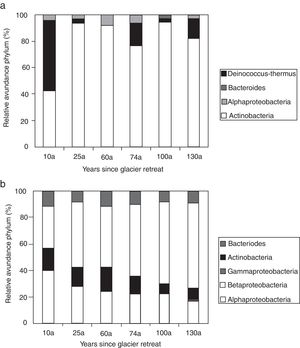
Actinobacteria was the dominant taxon for the samples incubated at 4°C, and the abundance remained constant along the chronosequence, whereas the abundance of Alphaproteobacteria and Bacteroides were relatively lower than Actinobacteria. The Deinococcus-Thermus group was only found in the 100a soil, and Betaproteobacteria and Gammaproteobacteria were not found in all of the samples (Fig. 3a). However, the species richness at 25°C was higher than at 4°C, including Actinobacteria, Bacteroides, and Alpha-, Beta- and Gammaproteobacteria. The Actinobacteria and Proteobacteria were the dominant taxa in the soils cultured at 25°C, with the abundance of Proteobacteria significantly decreasing along the chronosequence (p=0.02) and the abundance of Actinobacteria significantly increasing along the chronosequence (p=0.01). The abundance of Bacteroides was relatively stable in all of the soil samples. The Betaproteobacteria were only found in the oldest soils, and the abundance was very low compared to the other taxa (Fig. 3b).
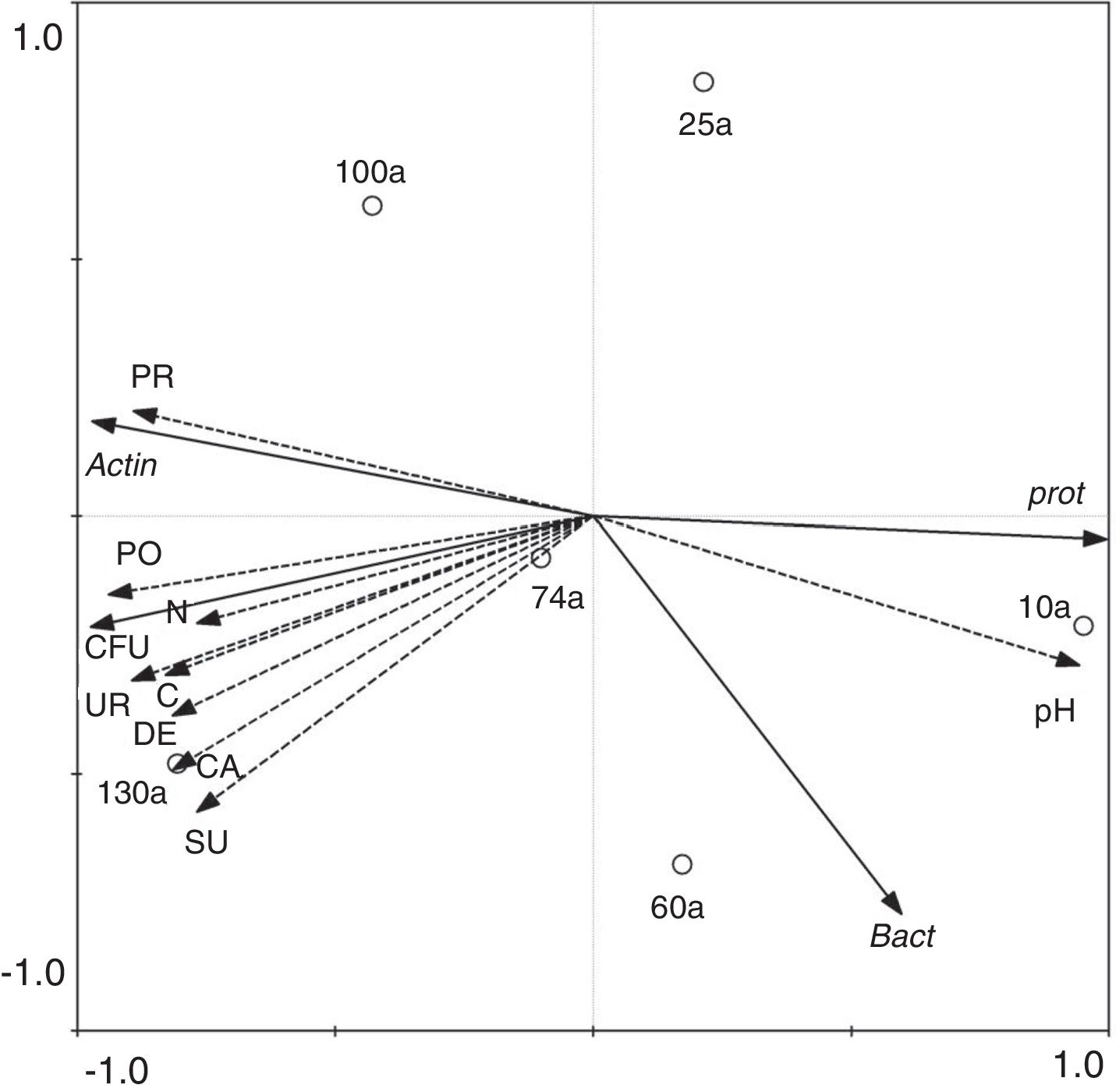
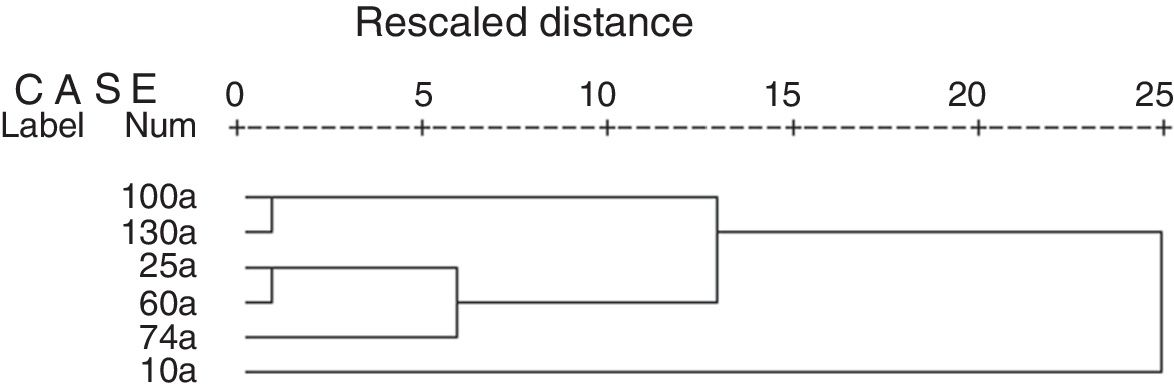
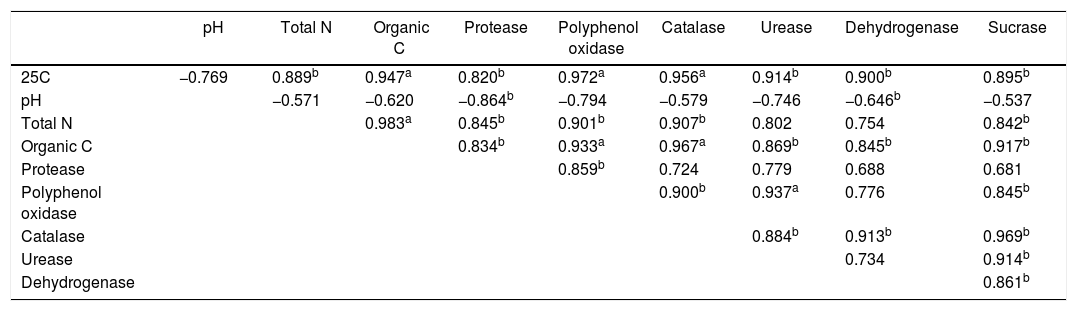
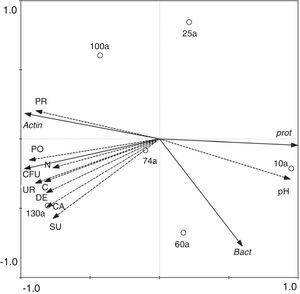
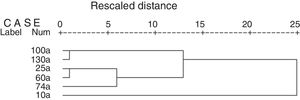
The correlations between the abundance of soil culturable bacteria and soil biochemical parametersThe total number of culturable bacteria in the soils cultured at 25°C were significantly positively correlated with the soil total N (p<0.05), organic C (p<0.01), and soil catalase (p<0.01), dehydrogenase (p<0.05), polyphenoloxidase (p<0.01), protease (p<0.05), sucrase (p<0.05), and urease (p<0.05) activities. The soil pH value negatively correlated with the number of culturable bacteria but was not significant. This result indicated that the bacterial abundance increased with increasing soil N and organic C and increasing soil enzyme activities and with decreasing soil pH values along the soil age gradient (Table 3). RDA axes 1 and 2 were found to explain 91.5% and 7.2%, respectively, of the overall variance, with the bacterial abundance and diversity data correlating with the environmental data. The number of culturable bacteria had a positive relationship with the soil enzyme activity, which was in good agreement with the SPSS correlation analysis. In addition, the bacterial communities of the 10a soils and 100–130a soils were separated from 25–74a, indicating that the bacterial succession in the Tianshan No. 1 glacier foreland should be divided into three stages: an early stage (10a), a middle stage (25–74a) and a late stage (100–130a) (Fig. 4). This result is consistent with the cluster analysis results (Fig. 5).
The correlations between the numbers of culturable bacteria at 25°C and the soil biochemical parameters along the chronosequences.
| pH | Total N | Organic C | Protease | Polyphenol oxidase | Catalase | Urease | Dehydrogenase | Sucrase | |
|---|---|---|---|---|---|---|---|---|---|
| 25C | −0.769 | 0.889b | 0.947a | 0.820b | 0.972a | 0.956a | 0.914b | 0.900b | 0.895b |
| pH | −0.571 | −0.620 | −0.864b | −0.794 | −0.579 | −0.746 | −0.646b | −0.537 | |
| Total N | 0.983a | 0.845b | 0.901b | 0.907b | 0.802 | 0.754 | 0.842b | ||
| Organic C | 0.834b | 0.933a | 0.967a | 0.869b | 0.845b | 0.917b | |||
| Protease | 0.859b | 0.724 | 0.779 | 0.688 | 0.681 | ||||
| Polyphenol oxidase | 0.900b | 0.937a | 0.776 | 0.845b | |||||
| Catalase | 0.884b | 0.913b | 0.969b | ||||||
| Urease | 0.734 | 0.914b | |||||||
| Dehydrogenase | 0.861b |
25C denotes the number of culturable bacteria at 25°C.
RDA biplot of the correlation between the taxa abundance of bacteria cultured at 25°C and soil biochemical parameters. The taxa abundance of bacteria cultured at 25°C (see Fig. 3b); soil biochemical parameters (see Table 2). Dash line arrows indicate the environmental variables (N, TN (%); C, OC (%); UR, urease; DE, dehydrogenase; SU, sucrase; CA, catalase; PO, polyphenol oxidase, PR, protease). Solid line arrows indicate the abundance of bacteria (Bact, bacteroides (%); Prot, proteobacteria (%); Actin, actinobacteria (%)).
The soil organic C, total N and enzyme activities increased along the exposure time gradient, while the soil pH decreased in the foreland of Tianshan No. 1 glacier. Similar results were reported at Dongkemadi glacier and Damma glacier.32,33 The total number of culturable bacteria recovered from the Tianshan No. 1 glacier foreland was higher than that of the permafrost in the same area (2.5–6.0×105CFUg−1)13 but similar to the number recovered from the Dongkemadi glacier foreland in the Central Tanggula Mountains (1.29×105–2.54×106CFUg−1).32 The soil organic C and total N have positive relationships with the soil enzyme activities and microbial abundance in the Tianshan No. 1 glacier foreland. Zumsteg et al.33 had findings similar to ours, reporting that the microbial community structure and enzymatic activity patterns are strongly conditioned by the successional stage in addition to the C and N contents of the glacier foreland soils. Furthermore, the incubation temperatures also affected the abundance and diversity of the bacteria. This finding is in accordance with the Lapanje et al.25 study at the Damma glacier and Liu et al.32 at the Dongkemadi glacier. The reason for the difference may be that, although they were not the dominant bacteria at the early age (10a), psychrotolerant bacteria become dominant with increasing soil age.34
The bacterial taxa and dominant phyla isolated from the Tianshan No. 1 glacier foreland were similar to the Qinghai-Tibet Plateau Jadang glacier foreland,35 Dongkemadi glacier foreland in the Central Tanggula Mountains,32 Himalayan Pindari glacier foreland,36 high Artic permafrost soil,37 and Antarctic permafrost soil.38 This result indicated that similar cold environments accommodate similar microbes, which could be evidence to support the “environment select” theory. Some of the bacterial isolates from the Tianshan No. 1 glacier foreland may originate from the glacier and glacier sediment, while others may derive from the atmospheric dust from other environments.39 The bacterial isolates from Tianshan No. 1 glacier foreland belong to six phyla. Bai et al.13 examined the phylogenetic diversity of bacteria from permafrost in the Tianshan Mountains using a culture-dependent method and found 4 phyla: Actinobacteria, Bacteroides, Firmicutes and Proteobacteria. Yang et al.14 studied the permafrost bacterial community structures and diversity using a denaturing gradient gel electrophoresis method and found 7 phyla of bacteria, including the Acidobacteria, Gemmatimonadetes and Chloroflexi phyla which were not found by Bai et al.13 Compared with the previous reports in this study area, the present study is the first to recover bacterial strains from the phylum Deinococcus-Thermus. Deinococcus-Thermus consists mainly of thermophiles,40 and finding Deinococcus-Thermus in this cold and higher UV exposure environment is interesting and should be further studied for its cold-adaption and UV-resistance mechanisms.
The dominate actinobacteria isolates in this study belonged to the genus Arthrobacter, which was the most highly diverse group among our isolates and are typically the predominant genus in cold environments.19,25,32,37,38,41 Proteobacteria are a favored taxon in this initial ecosystem with a low nutrient content, because many of them have a range of metabolism.42 Some of the genera in our study have been reported by others to have particular weathering capabilities, such as Sphingomonas sp.,43Pseudomonas sp.44 and Janthinobacterium sp.25 These genera should be further investigated for these capabilities. Studies show a positive correlation between Proteobacteria and the soil pH; in contrast, a negative relationship between Actinobacteria and soil pH was reported by Philippot et al.45
A previous study found that bacterial numbers depended mostly on the soil age after deglaciation in the Damma glacier foreland46 and in the Lyman glacier foreland.47 Liu et al.32 also found similar results. These results suggest that the number and activity of microbial populations increase with soil development after glacial retreat.46 Other studies also reported that microbial abundance has significant correlations with soil enzyme activities in the mountain valley wetland,48 reclaimed soil on a surface coal mine,49 and paddy soil.50 The RDA and hierarchical cluster analysis showed the same result, that the bacterial succession in the Tianshan No. 1 glacier can be divided into three successional periods: an early stage (10a), a middle stage (25–74a), and a late stage (100–130a). Similar results were reported by Huang et al.51 for copper mine tailings. These results indicate that soil microbes play a role in the development of the bare soil in the glacier foreland, suggesting that microbial succession correlates with the soil age along the foreland.
Conflicts of interestThe authors declare no conflicts of interest.
This work was funded by the Natural Science Foundation of China (Nos. 31500429, 31170465), the National Basic Research Program (973) of China (No. 2012CB026105), the China Postdoctoral Science Fund (2014M562477), and the Science and Technology Projects in Gansu Province (1506RJZA291) China.