In recent years, many studies have related gut microbiome to development of highly prevalent diseases such as type 2 diabetes and obesity. Obesity itself is associated to changes in the composition of gut microbiome, with a trend to an overgrowth of microorganisms more efficiently obtaining energy from diet. There are several mechanisms that relate microbiota to the onset of insulin resistance and diabetes, including changes in bowel permeability, endotoxemia, interaction with bile acids, changes in the proportion of brown adipose tissue, and effects associated to use of drugs like metformin.
Currently, use of pro and prebiotics and other new techniques such as gut microbiota transplant, or even antibiotic therapy, has been postulated to be useful tools to modulate the development of obesity and insulin resistance through the diet.
En los últimos años son muy numerosos los trabajos que han relacionado la microbiota intestinal con el desarrollo de enfermedades de alta prevalencia como son la diabetes tipo 2 y la obesidad. La obesidad por sí misma se asocia con cambios en la composición de la microbiota intestinal con tendencia al sobrecrecimiento de microorganismos con una mayor eficiencia en la obtención de la energía de la dieta. Son varios los mecanismos que relacionan la microbiota con la aparición de insulinorresistencia y diabetes, entre ellos destacan los cambios en la permeabilidad intestinal, endotoxemia, interrelación con ácidos biliares, cambios en la proporción de tejido adiposo marrón y efectos asociados al uso de fármacos como la metformina.
Actualmente, a través de la dieta, el uso de pro y prebióticos y otras nuevas técnicas como el trasplante de microbiota intestinal, o incluso la terapia con antibióticos, se postulan como herramientas útiles para modular la aparición de obesidad e insulinorresistencia.
Microorganisms living on and inside the human body form our microbiota, and their genes are known as microbiome.
Approximately 10–100 trillion microorganisms live in the adult bowel. They weight 1.5kg and are approximately 1000 species which exceed by 100 the human genome. The vast majority live in the colon.
Components of microbiota are mostly bacteria, with a minority of viruses, fungi, and eukaryotic cells. The most abundant phyla in both human and mice are Firmicutes, which account for 60–80% and include more than 200 genera (of which the most important are Ruminiococcus, Clostridium, and Lactobacillus); Bacteroidetes (particularly including Bacteroides, Prevotella, and Xylanibacter), accounting for 20–30%, and Actinobacteria, which represent a minority of approximately 10% (with predominance of the genus Bifidobacterium). Proteobacteria such as Escherichia and Enterobacteriaceae are less commonly found (Table 1). Microorganisms have significant interactions with each other and with the host.1
Gut microbiota is involved in a variety of metabolic functions such as fermentation and absorption of undigested carbohydrates, absorption of electrolytes and minerals, modulation of bowel motility, and synthesis of some micronutrients.2 Because of its role in performance of these functions, microbial changes in human bowel have been proposed as a potential cause of obesity.3
In addition to its metabolic functions, microbiota is involved in interaction with the immune system, providing signals to promote maturity of immune cells and normal performance of their functions, as well as toxin and carcinogen destruction, preventing colonization by pathogenic bacteria.2
Composition of gut microbiota depends on age, sex, geographical area, ethnicity, family, and diet, and may be modulated by prebiotics, probiotics, and antibiotics.4
Babies already acquire their initial microbiota at the time of delivery, especially from the maternal vagina or fecal microflora. By contrast, babies born by a cesarean section have a microbiota characteristic of the skin. These differences by type of delivery appear to influence the immunity developed in the first year of life, leading to different gut microbiota.5
Proteobacteria and Actinobacteria predominate in the first few days after delivery. Bacterial composition starts to coverage toward an adult profile of microbiota at the end of the first year of life, as the child grows and starts food intake. Microbiota increases in diversity and is completely similar to adult microbiota at approximately two and a half years of age. From this stage, Firmicutes and Bacteroidetes predominate.
During this time, the immune system “learns” to differentiate between commensal and pathogenic bacteria. Once microbiota has reached maturity, most of it remains stable until old age. The ELDERMET consortium studied microbiota in elderly people and found a characteristic composition different from that seen in young adults, particularly in the proportions of the Bacteroides and Clostridium groups.6,7
There has been an increasing interest in microbiota in recent years, which has resulted in an exponential increase in the number of publications on the subject. Attempts are being made to clarify the relationship of microbiota to development of highly prevalent diseases such as diabetes and obesity.8
It should be noted that microbiota is not an immutable entity, but may be modulated over time by changes in the environment and different influences. Changes in human ecology have affected the composition of microbiota during human evolution, but a more radical change has occurred in recent decades.
One of the most significant findings is that in developed countries there has been a loss of certain species that colonized our bowel some decades ago, with the resultant loss of biodiversity of our microbiota.
Factors that have influenced this change in microbiota include water sanitation, increased performance of cesarean sections, more frequent use of antibiotics in preterm newborns, decreased breast-feeding, the new model of small families, increased hygiene, or widespread use of antibacterial soaps.9
The microbiotas of European and African children have completely different compositions. African children have greater proportions of Bacteroidetes and Gram-positive organisms in their bowels, while a Western lifestyle appears to promote increases in Firmicutes and Gram-negative organisms.10
One of the most important factors that may alter composition of microbiota is use of antibiotics. Although the specific taxon affected varies depending on the subject, some taxa do not recover even after months of treatment, and there is usually a long-term decrease in bacterial biodiversity after use of antibiotics.
In relation with these assumptions, research on isolated populations has found a greater variety of microorganisms as compared to industrialized subjects.11
Other studies in humans at specific stages, such as pregnancy, have identified changes in maternal microbiota as an adaptive mechanism to the fetus and the different body composition.12,13
Methods to measure microbiotaIn order to ascertain the mechanisms that could be involved in development of obesity and other highly prevalent diseases, large scale projects have been undertaken, such as the Human Microbiome Project and MetaHIT.14,15
Research in this field is mainly performed using 16S ribosomal RNA (rRNA) and whole-genome shotgun, and has provided an overview of the commensal microbial communities and their functional capacity.14–16
These studies have shown a great variability in microbiota composition in healthy individuals. They have even shown that twins share less than 50% of their bacterial taxa at species level. This does not mean, however, that genetics does not play a role in the establishment and shaping of gut microbiota, as composition of the bacterial community has been shown to be influenced by host-specific genomic loci.17,18
Traditionally, study of gut microbiota has mainly been based on culture of microorganisms and in their identification through conventional morphological, physiological, and chemical phenotypical tests.
Culture of fecal microbiota in selective, differential media appears to be the simplest, most direct method; it is however poorly reliable because some bacteria cannot be cultured. Ninety-nine percent of bacteria in fecal contents are strict anaerobes, and many of them are extremely sensitive to oxygen. Strict reducing conditions are therefore required during processing and culture.
Indirect methods basically consist of the study of bacterial metabolism. The principle is based on estimation of gut microbiota by testing and quantification of their metabolites or some enzymatic activities. Metabolites such as volatile fatty acids or products of bile acid metabolism may be tested using chromatographic techniques, or enzymatic activities of microbial origin may be analyzed. These study methods have the disadvantage that many enzyme activities are not specific of a particular microorganism or bacterial group. In addition, species sometimes have a great metabolic variability or plasticity.
The disadvantages of traditional study methods have led to develop alternative approaches. Molecular genetics tools other than culture have a great potential for identification, quantification, and typing of gastrointestinal tract microorganisms. Polymerase chain reaction, with all its variants, is of the most widely used procedures to estimate non-culturable microorganisms. Many approaches are also based on differences in the sequence of the gene that encodes 16S rRNA, which because of the alternation of conserved and variable regions, with clear phylogenetic implications, is particularly useful for studying microbial diversity.
16S rRNA is the macromolecule most widely used in bacterial phylogeny and taxonomy studies.19
The most commonly used procedures include:
- –
Fluorescence in situ hybridization. This quantitative procedure uses rRNA probes marked with fluorescence that directly hybridize with bacterial preparations fixed on a slide. Fluorescence microscopy is used for detection or visualization.20
- –
Construction of genomic libraries of 16S rDNA sequences obtained by direct amplification of bacterial DNA from the samples. If amplification is not biased, the number and diversity of clones in the gene library will reflect the species present in the original sample. Microbial diversity is determined after sequencing and comparison of sequences with those in the databases.21
- –
Denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis. Both methods are based on separation of amplification fragments with different sequence using electrophoresis in a chemical denaturing or temperature gradient.22
- –
Gnotobiosis is an in vivo method to study gut microbiota using germ-free experimental animals. After delivery, usually by cesarean sections, animals are directly placed in sterile cabins where, in addition to atmosphere, all materials and nutrients provided are sterile. In other cases, sterility is achieved with a first bottle containing antibiotics that makes bacteria implantation in the gastrointestinal tract impossible. In these animals, microorganisms may subsequently be introduced under controlled conditions for their study. Many of the roles attributed to gut microbiota have been determined by comparing axenic (sterile) animals and holoxenic animals (conventional animals) with normal microbiota.23
Obesity is a pandemic condition associated to multiple metabolic changes and involving several organs and systems.
During the past decade, several studies have reported a causal relation between gut microbiota and development of metabolic diseases such as diabetes and obesity. A new paradigm suggesting that microbiota may contribute to regulation of energy homeostasis emerges.
It is hypothesized that interrelation of environmental circumstances with gut microbiota may cause an energy imbalance leading to metabolic, neurocognitive, and behavioral changes that would promote development of obesity.24–26
Independent contribution of microbiota to fat accumulation has been shown in a number of in vivo studies in mice. Germ-free mice, having no microbiota, have significantly less body fat than normal mice despite eating more.27
The experiment most strongly supporting a causal relationship between microbiota and obesity probably was the one conducted by Turnbaugh et al. in 2006, which showed that transplantation of microbiota from genetically obese to germ-free mice caused a very significant weight increase as compared to germ-free mice that were transplanted microbiota from thin mice.28
Studies in mice have found a greater abundance of Firmicutes in obese mice and in those fed Western diets, concomitant with a decreased abundance of Bacteroidetes.3,29 Among Firmicutes phyla, the class Mollicutes was most common in obese mice.29
The increase in Firmicutes seen in animals, and also in obese subjects, could be associated to an increased capacity for digesting some indigestible polysaccharides to monosaccharides and short-chain fatty acids that may be absorbed by the host, thus obtaining more energy from substances which in thin subjects would be excreted in feces unabsorbed.3,30,31
Bacteroidetes have fewer genes for enzymes involved in lipid and carbohydrate metabolism than Firmicutes.32 However, within the Bacteroidetes phylum, Bacteroides thetaiotaomicron has been shown to improve nutrient absorption and processing by the host.33
Studies in humans reported disparate results. Some supported the finding of a high Firmicutes/Bacteroidetes ratio,8,34–36 others found no correlation between body mass index and Firmicutes/Bacteroidetes ratio,37,38 and still others found an opposite ratio.30,39
Obese patients included in a low-calorie diet program or those undergoing Roux-en-Y gastric bypass had decreased proportions of Firmicutes and/or increased proportions of Bacteriodetes.40–42
An additional important finding in humans was that switching from a fat-rich, low-fiber diet to a low-fat, fiber-rich diet induced significant changes in gut microbiota intestinal in only 24h. Moreover, multiple evidence suggests that, in humans, an increase in dietary fat decreases the number of Lactobacillus organisms and causes an increase in Gram-negative bacteria.43,44
Specifically, a higher level of Lactobacillus reuteri and lower levels of Lactobacillus casei/paracasei and Lactobacillus plantarum were associated to obesity.45
Zhang et al. suggested that increased energy storage in obese individuals is related to hydrogen transfer between taxa, because they noted a simultaneous increase in hydrogen-producing Prevotella and methanogenic Archaea using nitrogen.40,45,46 Methanogenic Archaea is able to convert hydrogen into methane and to obtain more energy from the same daily calorie intake.47,48
On the other hand, gut microbiota may decrease production of fasting-induced adipocyte factor by intestinal cells, thus inhibiting activity of the enzyme lipoprotein lipase. This enzyme promotes release of non-esterified fatty acids to tissues such as the liver and adipose cells.49
Several studies have investigated the association of given species to human obesity. In children and pregnant women, an association was shown between Staphylococcus aureus and overweight.39,50 Lower numbers of Bacteroides and greater numbers of Staphylococcus, Enterobacteriaceae, and Escherichia coli (E. coli) have been reported in overweight subjects as compared to pregnant women of normal weight.36 Levels of Faecalibacterium prausnitzii (F. prausnitzii) (from the phylum Firmicutes) were significantly higher in obese as compared to non-obese children.51 The proportions of the Bacteroides-Prevotella group were shown to increase after weight loss in obese adolescents.52
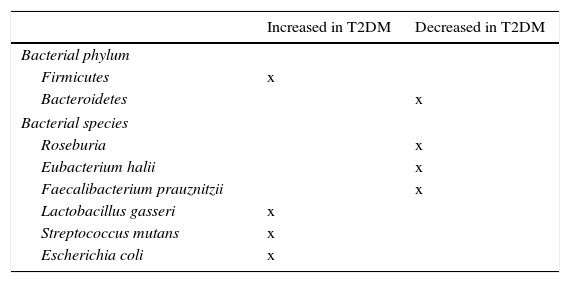
Role of microbiota in diabetes developmentMassive pyrosequencing procedures have allowed for substantial advances in characterization of our microbiota. As compared to non-diabetic subjects, patients with diabetes had a decrease in butyrate-producing bacteria such as Roseburia intestinalis and F. prausnitzii, and increases in Lactobacillus gasseri (L. gasseri), Streptococcus mutans (S. mutans), and some Clostridium microorganisms (Table 2). A greater proportion of Proteobacteria and increased expression of microbiota genes involved in oxidative stress and inflammation.53,54
Bacterial species related to occurrence of insulin resistance and type 2 diabetes mellitus (T2DM).
| Increased in T2DM | Decreased in T2DM | |
|---|---|---|
| Bacterial phylum | ||
| Firmicutes | x | |
| Bacteroidetes | x | |
| Bacterial species | ||
| Roseburia | x | |
| Eubacterium halii | x | |
| Faecalibacterium prauznitzii | x | |
| Lactobacillus gasseri | x | |
| Streptococcus mutans | x | |
| Escherichia coli | x | |
In studies conducted by our group comparing metabolically healthy and insulin-resistant obese subjects, significant changes associated to insulin resistance continue to be seen.55
Several mechanisms have been proposed to explain the impact of microbiota on insulin resistance. The mechanisms most strongly supported in the literature are discussed below.
Increase in endotoxemiaType 2 diabetes mellitus is associated to a pro-inflammatory state with a moderate excess production of cytokines such as IL-6, IL-1, or tumor necrosis factor-α, which impair insulin interaction with its receptor and contribute to insulin resistance and diabetes. Weight increase appears to be one of the factors triggering this low-grade inflammation.
Animal experiments have shown that changes in la microbiota may change the grade of adipose tissue inflammation. Lipopolysaccharides (LPS) are components of the cell wall of Gram-negative bacteria. Increased circulating LPS levels have been found in subjects with greater fat intake.56,57 LPS are absorbed by enterocytes and carried in plasma mainly bound to chylomicrons.58
The causative role of LPS has been shown, as infusion of LPS into mice fed a normal diet induced insulin resistance in the liver, glucose tolerance, and increased adipose tissue weight. LPS binds to the CD14/TLR4 receptor in macrophages and induces an increase in production of pro-inflammatory molecules. When LPS injections were administered to mice with a genetic absence of the CD14/TLR4 receptor, mice did not develop these metabolic characteristics and did not experience type 2 diabetes or obesity, which shows the significant role of the mechanism of the CD14/TLR4 receptor for LPS. In addition, knockout CD14/TLR4 mice were even more sensitive to insulin than wild type controls.59
This suggests that in certain situations, a change occurs in the proportion of Gram-negative bacteria in the bowel, or there is a change in intestinal permeability so that LPS increase in serum, and this increase is directly related to the degree of insulin resistance.
Changes in incretin secretion related to insulin resistance and beta cell functionIt has been shown that increases in Bifidobacterium spp. modulate inflammation in obese mice by increasing production of glucagon-like peptide-1 (GLP-1), also reducing intestinal permeability. There is evidence that the increase in Bifidobacterium spp. induced by some prebiotics is associated to increased secretion of GLP-1 and peptide YY by the bowel; these two molecules have favorable effects, decreasing insulin resistance and increasing beta cell function.60
Changes in butyrate productionButyrate is a short-chain fatty acid (SCFA) which, together with propionate and acetate, are produced by gut bacteria by digesting fiber.61
SCFAs are absorbed in the bowel, where they provide (especially butyrate) energy to colonic epithelial cells, while the rest enter the portal venous system and butyrate contributes very significantly to decrease intestinal permeability.
Data from animal studies suggest that propionate affects lipogenesis and gluconeogenesis in the liver, while the only role of acetate at peripheral level is to act as a substrate for the synthesis of cholesterol.62
Interestingly, butyrate production has been shown to affect serotonin levels in animal models. And directly affects sympathetic tone, bowel transit time, and physical activity.63
It is now recognized that serotonin may regulate intestinal permeability, in addition to being an important neurotransmitter in the bowel and brain involved in body weight regulation and food intake by controlling satiety. Reductions in brain production of SERT, essential regulators of serotonergic transmission, is associated to obesity.64
Changes in characteristics of brown adipose tissueObesity is characterized by a reduction in thermogenic activity of brown adipose tissue (BAT). BAT promotes a thin, healthy phenotype and improves insulin sensitivity. In response to cold or exercise, brown fat cells also arise in white adipose tissue (WAT), where they are also known as beige fat cells. This process is called “browning”.
Development of beige fat in subcutaneous or visceral adipose tissue is achieved in animal models after broad spectrum antibiotic treatment that eradicates microbiota or in germ-free mice. This leads to improved glucose tolerance and insulin sensitivity, and to decreases in white fat and adipocyte size in mice.
These effects are reversed by recolonization with microbes of antibiotic-treated or germ-free mice.65
A recent study revealed that gut microbiota hinders the occurrence of brown adipocytes, known as beige adipocytes, included in regular WAT through a mechanism that involves control of macrophages and eosinophil infiltration.66
Impact of secondary bile acidsA majority of conjugated primary bile acids are reabsorbed through the enterohepatic circulation, and only 5% escape this mechanism and reach the large bowel, where they are converted into secondary bile acids mainly by the action of Firmicutes.
Lower numbers of secondary bile acids were seen in subjects with overweight and type 2 diabetes as compared to healthy subjects. This appeared to be more related to an impaired carbohydrate metabolism than to obesity.
Secondary bile acids appear to have an insulin-sensitizing role. They act as mediator molecules through nuclear receptors such as the FXR and the membrane receptor TGR5 expressed in various tissues, including gallbladder, ileum, colon, BAT, and WAT.67
In BAT and muscle, it increases mitochondrial activity and phosphorylation, and leads to insulin sensitization in models of diabetic and obese mice. On intestinal L-cells, it appears to improve glycemic metabolism, stimulating production of peptides such as GLP-1 and promoting insulin secretion.
The role of choline and niacin as vitaminsFirmicutes, Actinobacteria, and Proteobacteria can break down choline. Their end-products have been associated to development of cardiovascular disease and diabetes, as they promote development of oxidative stress. Similar results have been found for niacin and end products of its metabolism with F. prausnitzii.67
Impact of drug treatmentAlthough it is clear that public health has substantially benefited from discovery of antibiotics, their widespread use is starting to pose health problems. In addition to occurrence of antibiotic resistance, these drugs may be potentially associated to the obesity epidemic.68
Treatment with metformin, common in patients with type 2 diabetes mellitus, also appears to be a potential modifier of gut microbiota in some studies, probably because of its gastrointestinal effects.69
Changes in gut microbiotaThere is a place for hope as regards the value of microbiota and its adaptation to achieve benefits for health. Through diet, using prebiotics and probiotics, antibiotics and novel procedures such as gut microbiota transplantation, encouraging results are apparently achieved.
As discussed above, diet changes composition of microbiota and metagenome expression irrespective of host genome.23,24 Recent studies using the Mediterranean diet have also provided relevant information on its benefits by modifying gut microbiota in obese subjects to prevent development of type 2 diabetes mellitus.70
When obese subjects were given a low-calorie diet, low in fat or carbohydrates, abundance of Bacteroidetes increased, while Firmicutes decreased.71,72
Consumption of probiotics and prebiotics has been shown to modify gut microbiota and to improve carbohydrate metabolism.73 Bacteria fermenting carbohydrates such as Bifidobacterium and Lactobacillus have been given as part of prebiotic treatment in studies with populations of different age groups.71
Along these same lines, when mice on a high-fat diet were fed prebiotics containing oligofructose, this restored levels of bifidobacteria and decreased endotoxemia while improving glucose tolerance.74.
Another study by our group showed that red wine consumption may significantly modulate growth of intestinal flora in humans, increasing the numbers of Enterococcus, Prevotella, Bacteroides, Bifidobacterium, Bacteroides uniformis, Eggerthella lenta, and Blautia coccoides-Eubacterium rectale and decreasing the amount of LPS. This suggests potential prebiotic benefits in the diet through red wine polyphenols.75,76
An additional line of intervention would be use of probiotics as nutritional supplements enriched with live bacterial strains, including Bifidobacterium and Lactobacillus species, which are able to alter intestinal flora so that it is beneficial for those who receive them. In mice, an antidiabetic effect has been shown after administration of probiotics containing certain strains of Lactobacillus, with a concomitant reduction in endotoxemia.77–79
Treatment with antibiotics (ampicillin plus neomycin) of genetically obese mice fed a fat-rich diet modified their microbiota and decreased insulin resistance and weight of the animals. In addition, animals given antibiotic treatment showed a surprising decrease in the degree of inflammation, oxidative stress, and macrophage infiltration in adipose tissue.80,81
Other studies, such as the one recently published by our group, relate improvement in carbohydrate metabolism to eradication of Helicobacter pylori.82
Gut microbiota transplantation was initially used in patients who developed pseudomembranous colitis after Clostridium difficile infection treated with antibiotics, in whom fecal material transplanted from healthy donors appeared to restore microbial balance in the bowel by replacing the more pathogenic intestinal bacterial strains by other more beneficial strains.83 The procedure has subsequently been extended to other gastrointestinal diseases, which has opened a new way for its use in diseases such as obesity, diabetes, and cardiovascular diseases.84–86
Some recently reported studies on the subject used males with insulin resistance and metabolic syndrome, who received an autologous fecal microbiota transplant, or an allogenic transplant from thin donors.85 Subjects who received transplants from thin donors showed a significant improvement in peripheral sensitivity to insulin. There were also increases in microbial diversity in the bowel and butyrate-producing bacteria.
An additional study in the Swedish cohort showed changes in gut microbiota, with greater concentrations of L. gasseri and S. mutans (both living in proximal bowel), as well as E. coli, which could help predict the possibility of insulin resistance development in postmenopausal women.86
These were small scale studies, and their results have not been reproduced by other groups, but have stimulated development of better procedures for identifying gut microbiota and its potential properties.
ConclusionsIdentification of gut microbiota related to obesity and type 2 diabetes has served as a stimulus for exponential advances in scientific production in recent years. There are multiple factors involved in changes in gut microbiota and its relationship to type 2 diabetes. Multiple possibilities are now available to change gut microbiota to our benefit, and are providing encouraging results.
Conflicts of interestThe authors state that they have no conflicts of interest.
Please cite this article as: Muñoz-Garach A, Diaz-Perdigones C, Tinahones FJ. Microbiota y diabetes mellitus tipo 2. Endocrinol Nutr. 2016;63:560–568.